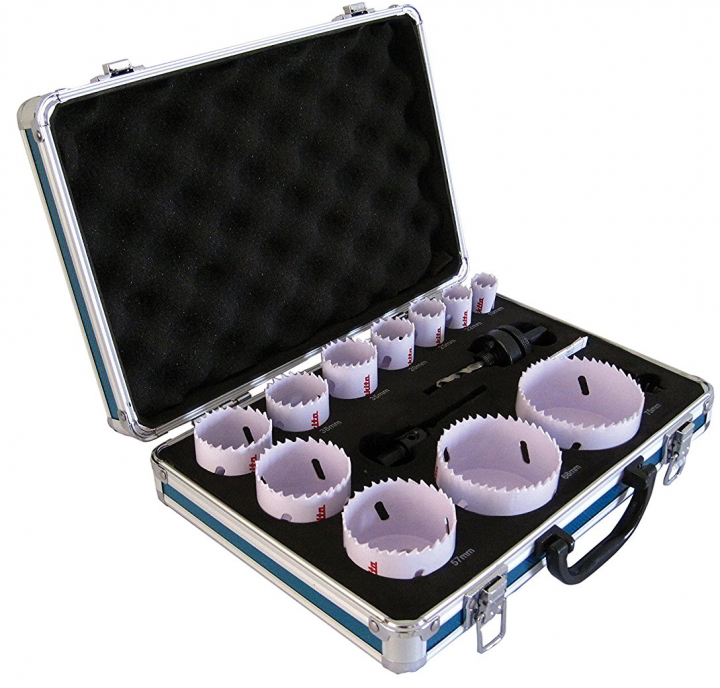
Vásárlás: Makita D-63965 körkivágó készlet 9 részes (D-63965) Körkivágó árak összehasonlítása, D 63965 körkivágó készlet 9 részes D 63965 boltok

Vásárlás: Makita Ezychange Bi-metal körkivágó készlet 7db-os (B-11972) Körkivágó árak összehasonlítása, Ezychange Bi metal körkivágó készlet 7 db os B 11972 boltok

Vásárlás: Makita HSS-BiCobalt körkivágó 102mm (P-52738) Körkivágó árak összehasonlítása, HSS BiCobalt körkivágó 102 mm P 52738 boltok

Makita Bimetál körkivágó 65mm (D-24882) | Aktív Kft. webáruház – Szerszám, fűtés, épületgépészet egy helyen

Makita Ezychange bimetál körkivágó 60 mm (B-11427) | Aktív Kft. webáruház – Szerszám, fűtés, épületgépészet egy helyen

Makita Ezychange körkivágó HSS-Bimetal 16mm | Tartozék | Körkivágó | Ezychange gyorscsatlakozós szerszámbefogás