
White Self-driving Passenger Drone Flying In The Sky. 3D Rendering Image. Stock Photo, Picture and Royalty Free Image. Image 88292576.
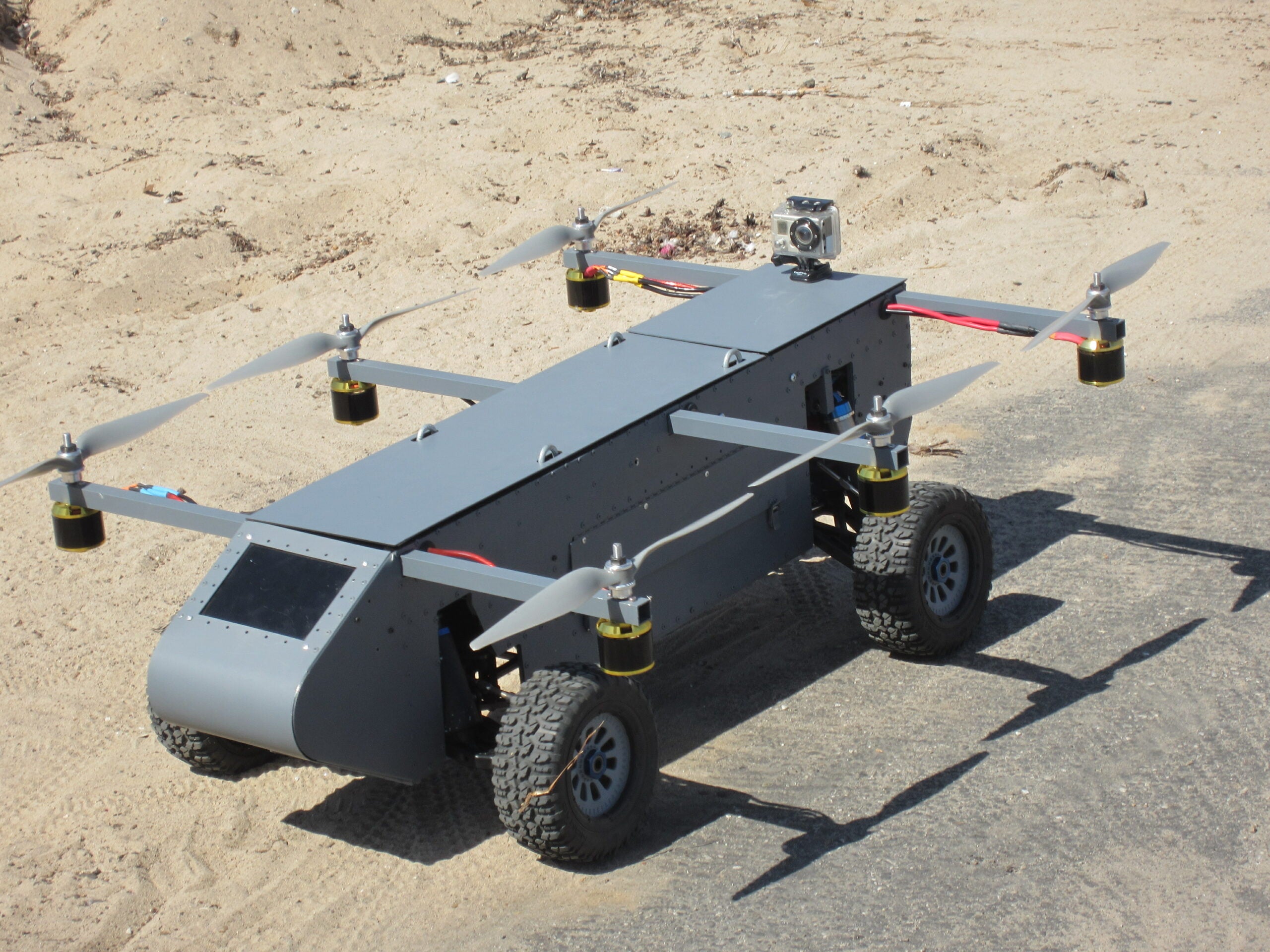
Top Row: A "Drivocopter" Drone (developed by the authors) which can fly... | Download Scientific Diagram

Sharper Image Fly+Drive Drone Remote Control Drone, Dual-Function Vehicle, White, 7 in. - Walmart.com

Sharper Image Fly+Drive Drone Remote Control Drone, Dual-Function Vehicle, White, 7 in. - Walmart.com




















/cdn.vox-cdn.com/uploads/chorus_image/image/63708594/img_6271.0.1484264832.0.jpg)


