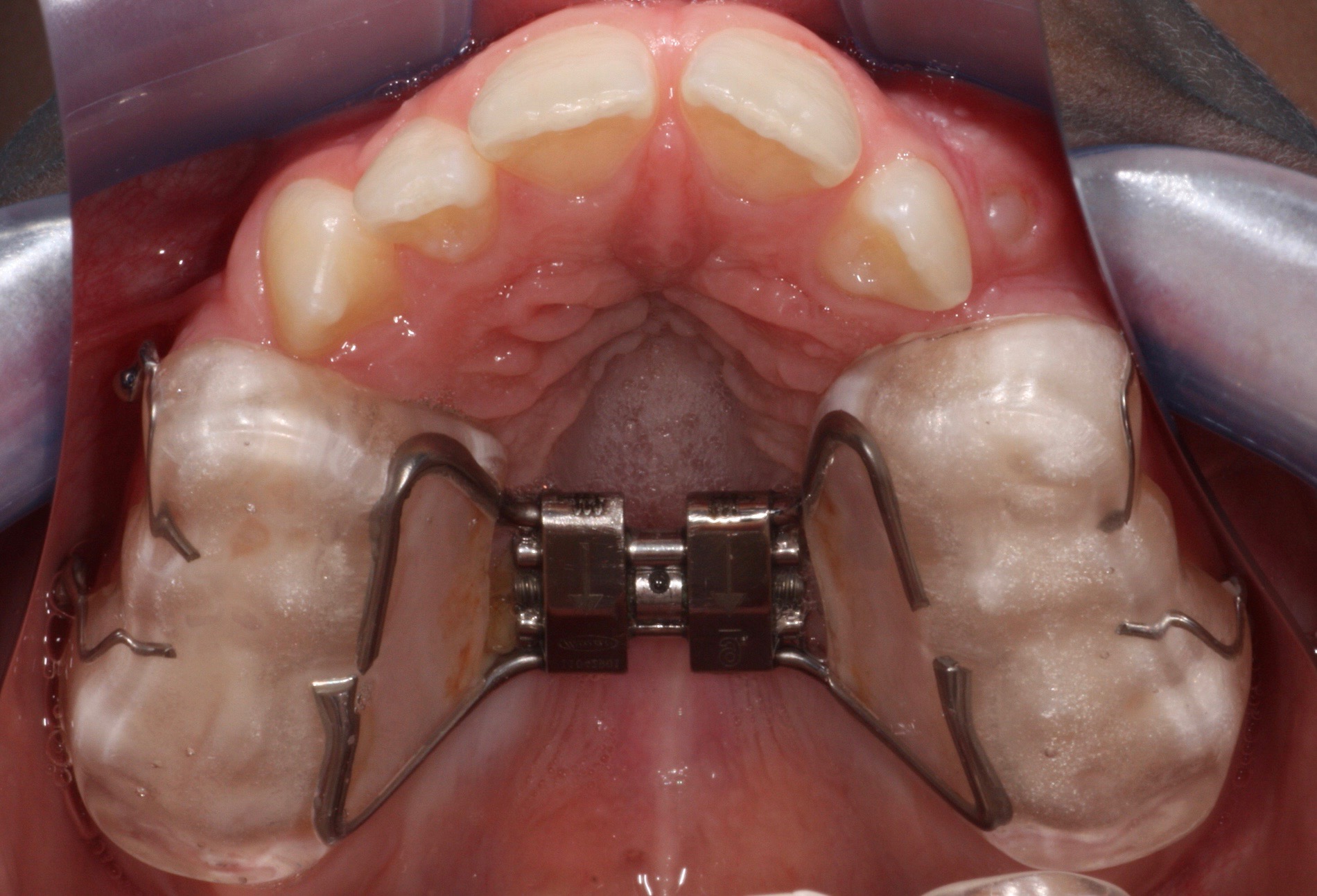
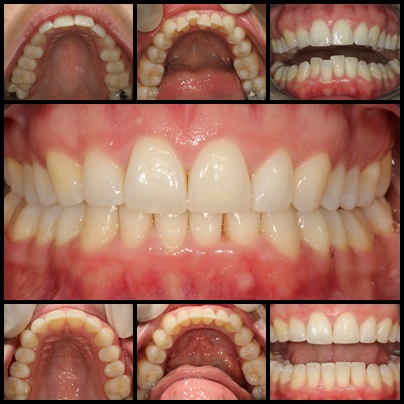
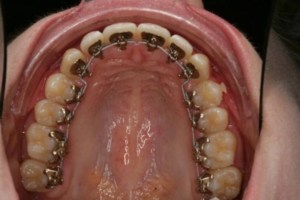
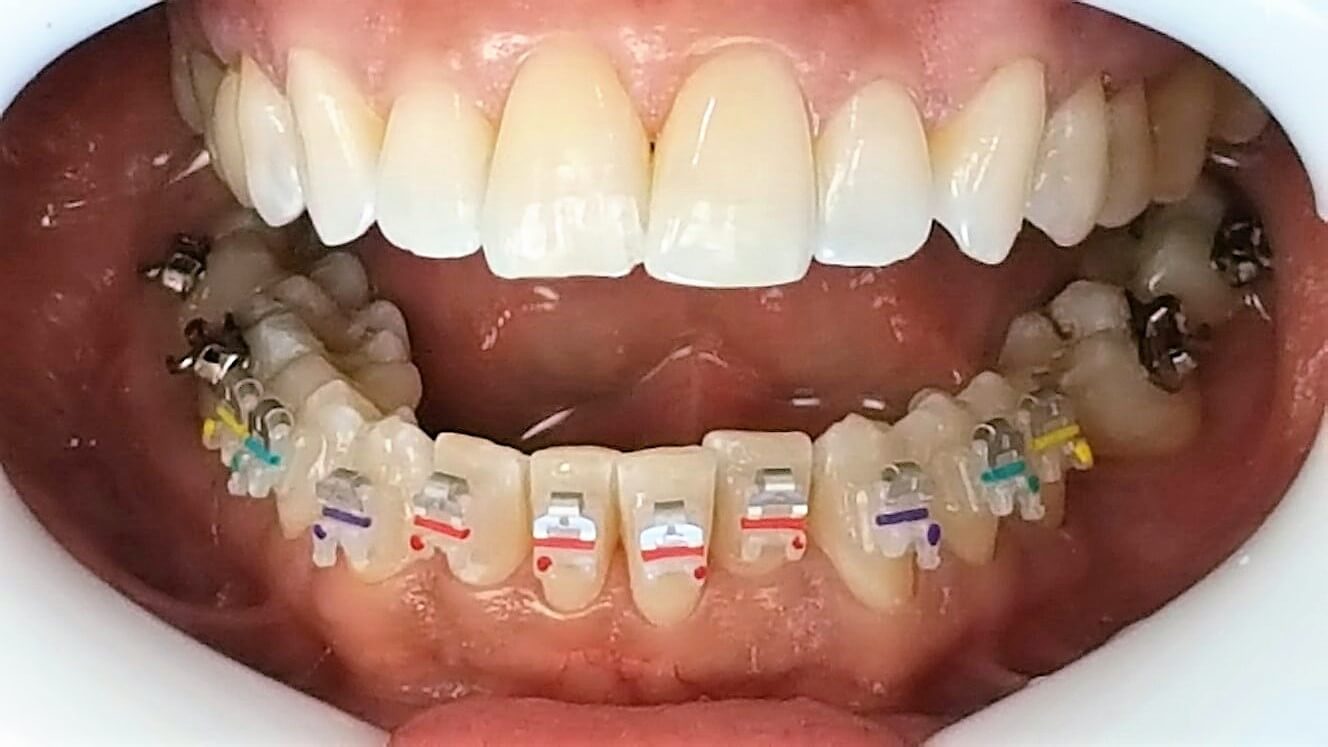
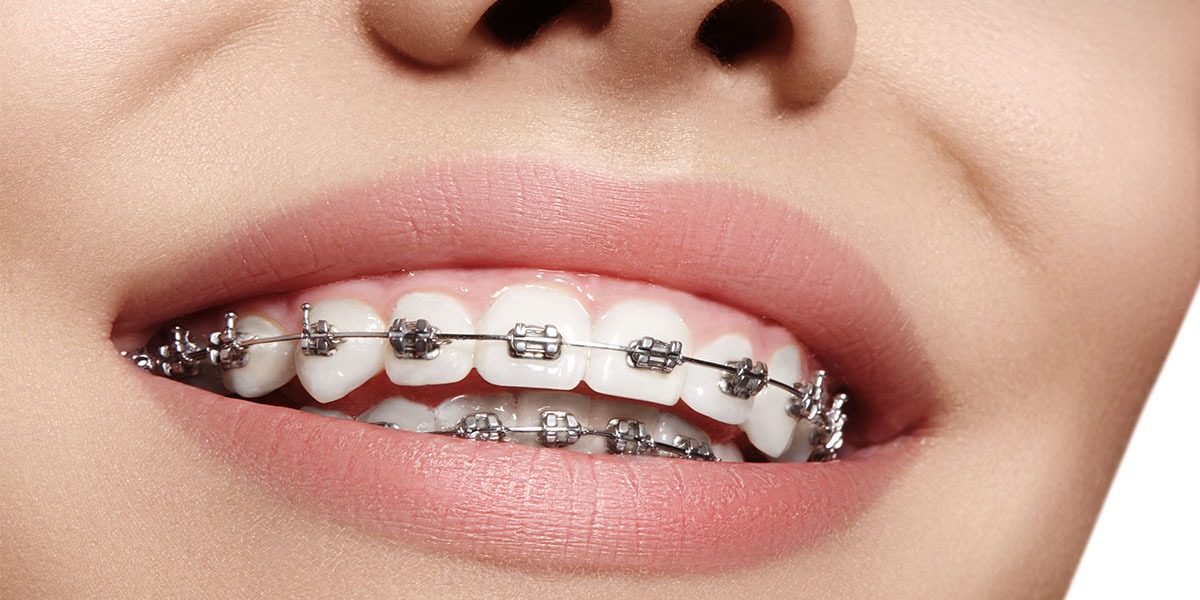
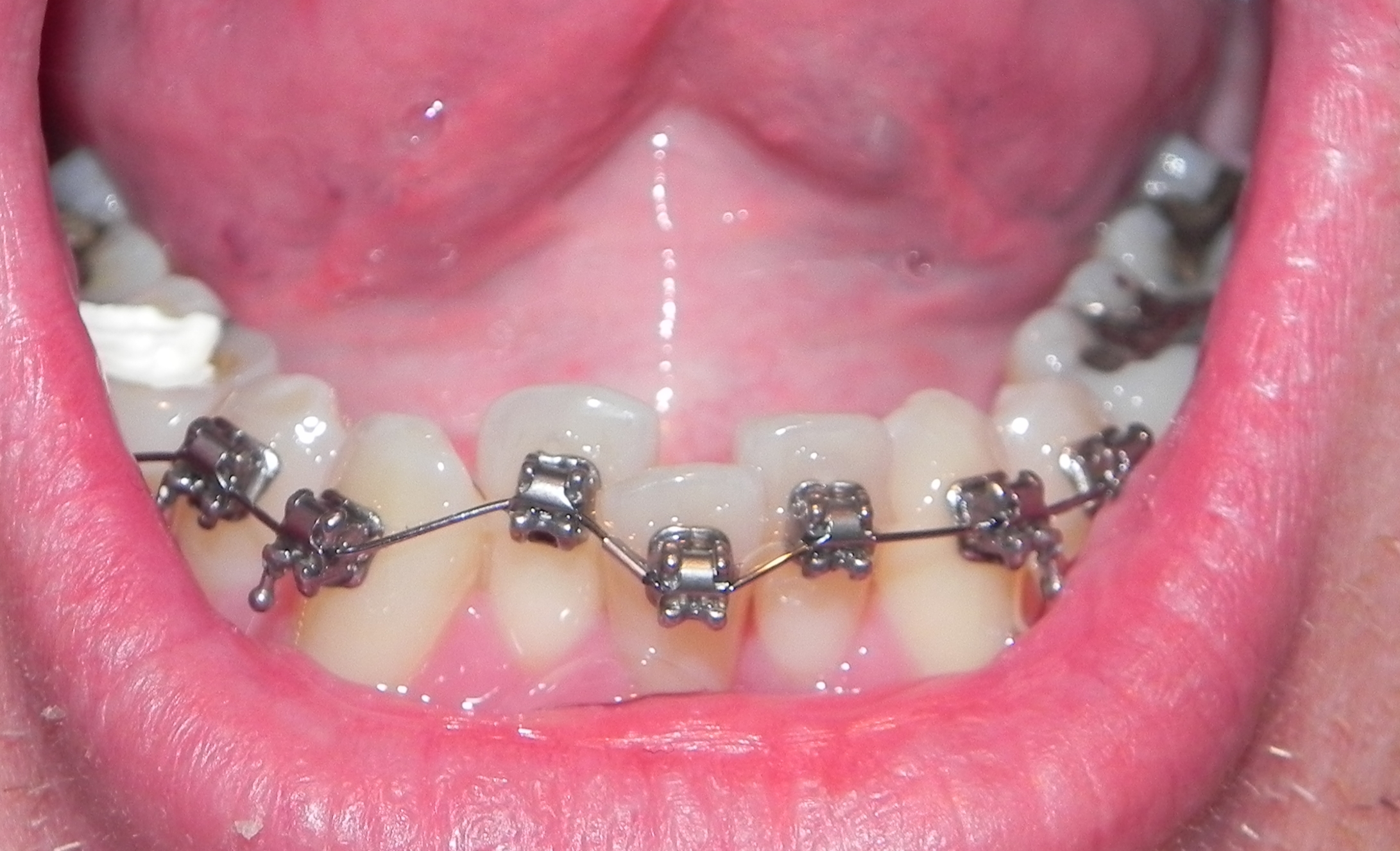
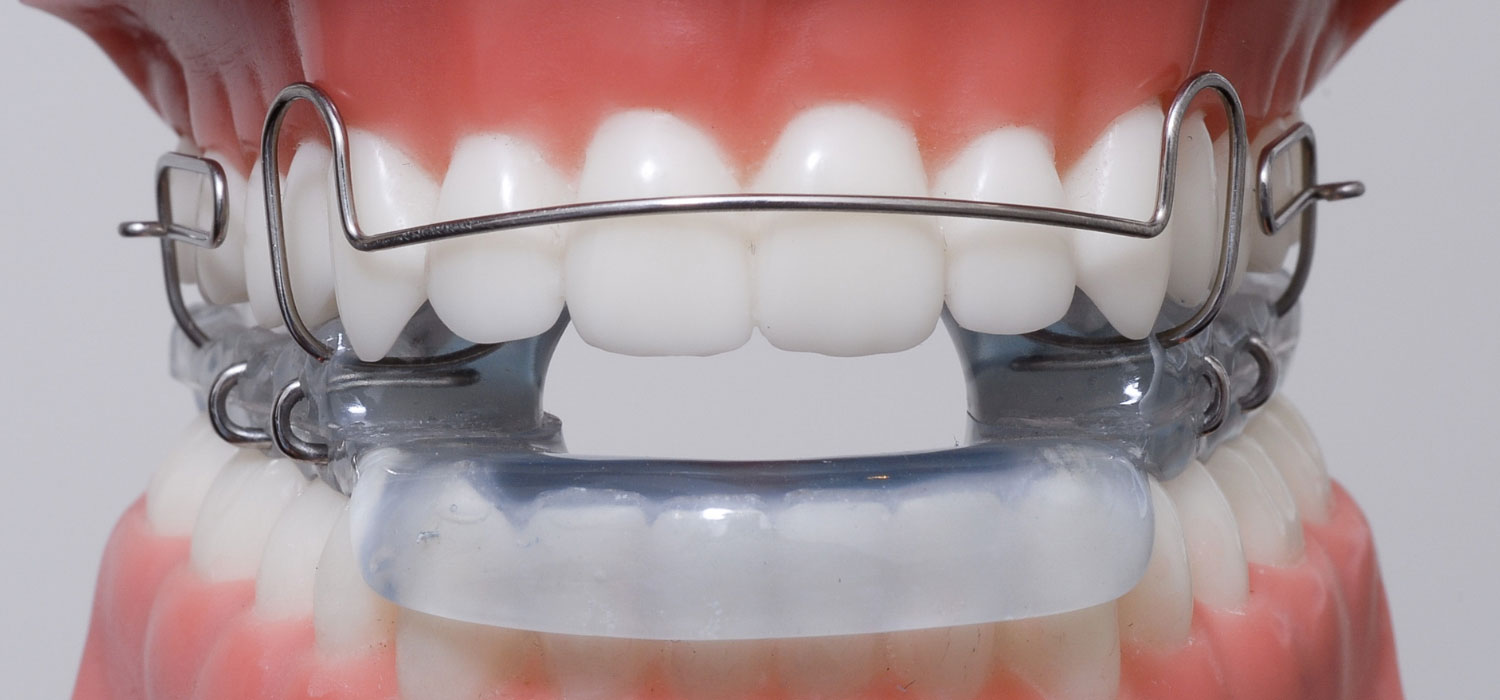
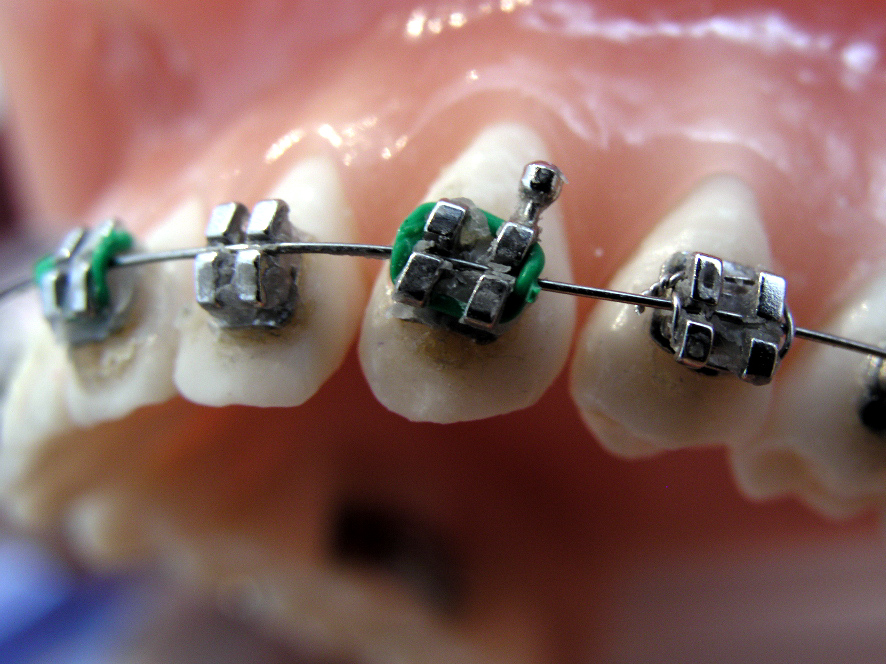
Orthodontie Eén van de specialisaties van tandarts Fabels is orthodontie. Bij orthodontie houdt de tandarts zich bezig met het optimaliseren van de stand van tanden in de kaken door middel van beugels. In het dagelijks leven is orthodontie te zien door de ...