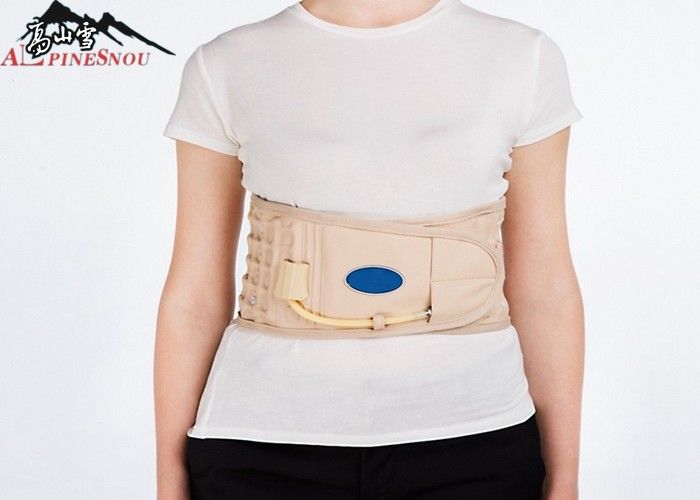
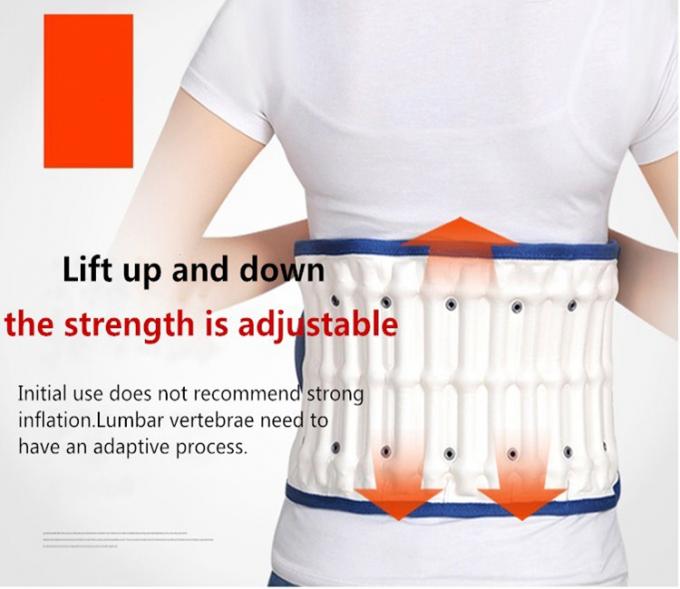
حزام الظهر، حزام سحب هواء العمود الفقري لتخفيف آلام أسفل الظهر، ودعم الظهر، حزام للجر أسفل الظهر : Amazon.ae: الجمال

حزام تصحيح للظهر المقوس غير المرئي لتحسين وضعية الجلوس وتقويم عظام الأطفال والبالغين., ابيض, : Amazon.ae: الصحة

حزام الظهر الطبي - قم بنشاطك الروتيني بدون اي ألم عن طريق حزام الظهر 😍 وخد بالك ده الحرام الاصلي 😎 السعر 199 جنيه والتوصيل مجانا 💪 حزام ﺍﻟﻈﻬﺮ ﺍﻟﻄﺒﻲ ﻫﻮ ﺣﺰﺍﻡ