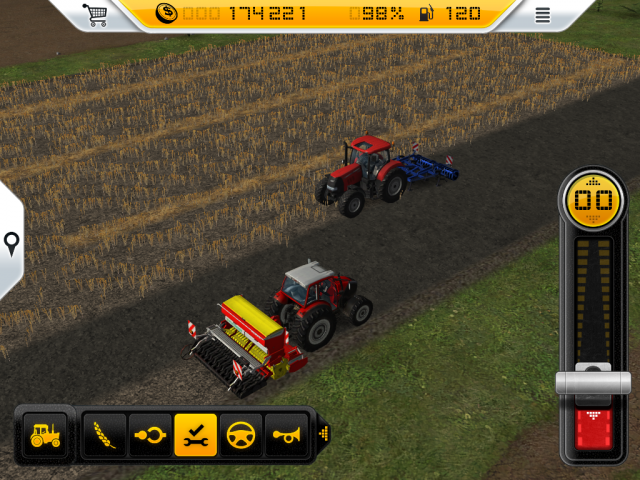
Farming Simulator - Farming Simulator 14 – Major update with three new vehicles and more equipment! Today we are releasing an update for Farming Simulator 14 on iOS and Android. It adds

Farming Simulator 14 review – one for virtual mucking out devotees | Simulation games | The Guardian

Farm Simulator 14 official release date announced, coming to Android on November 18th - Droid Gamers









:format(jpeg)/cdn.vox-cdn.com/uploads/chorus_image/image/33604303/farming_simulator_14-02.0.jpg)