
Amazon.com | Susanny Women Autumn Round Toe Lace Up Ankle Buckle Chunky Drag High Heel Platform Knight Black Martin Boots 5.5 B (M) US (CN Size_37) | Ankle & Bootie
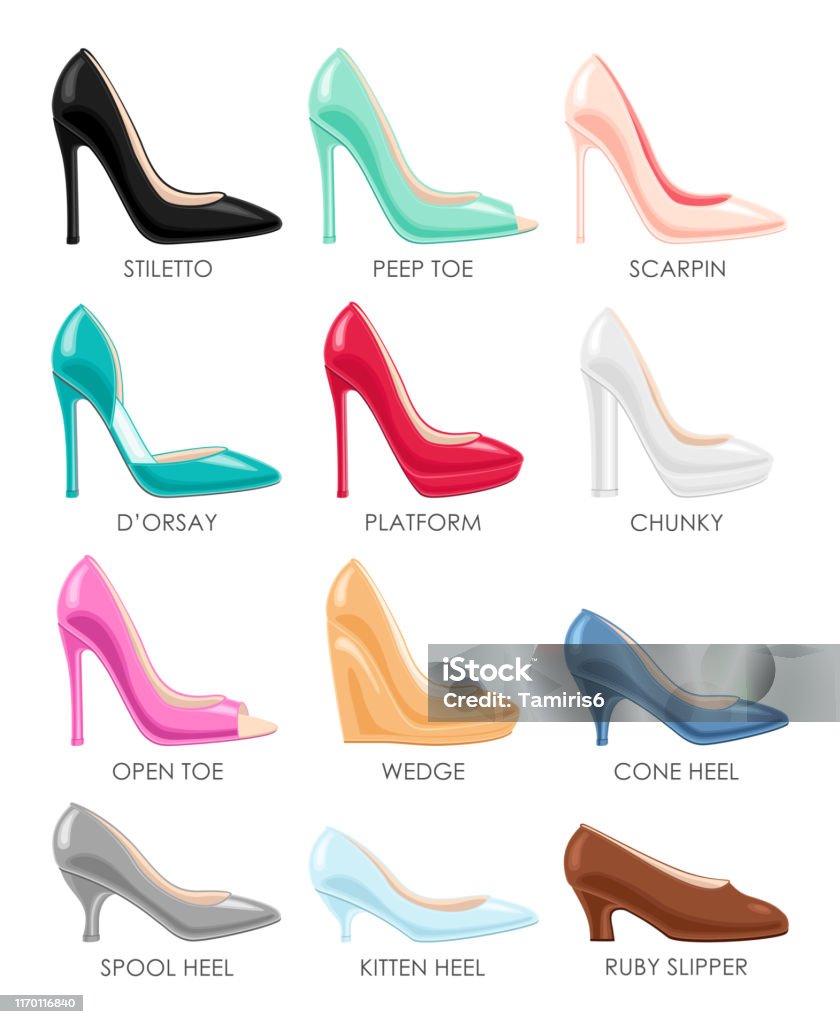
Set Of Different Types Of Female High Heeled Platform Wedge Stiletto Shoes Fashion Background Stock Illustration - Download Image Now - iStock

Amazon.com | Aachcol Women Platform Pumps Stiletto High Heel Close Round Toe Slip-on Dress Shoes Classic 6 Inch Beige Natural Patent 5 M US | Shoes

High heel shoes - Women's sizes 6-11 - Clear platform 6 inch stiletto heel - Black patent sandal and straps | Enjoyables By JR

Silver shiny Giaro SLICK ESCALA platform pumps with metal heels - Giaro High Heels | Official store - All Vegan High Heels


















