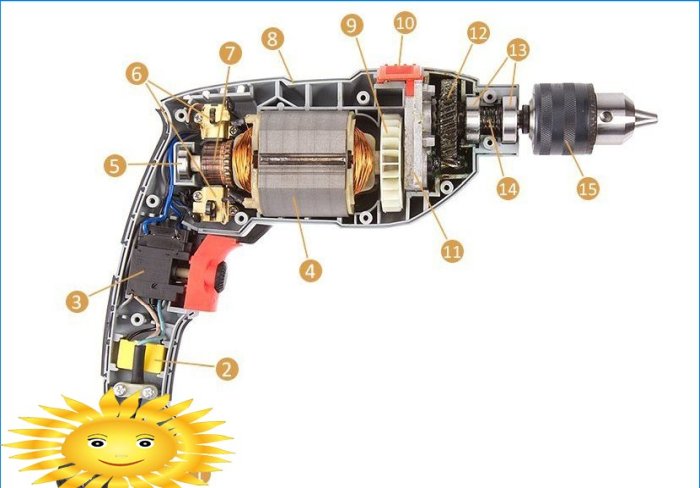
Zamjena i popravak sidra brusilice vlastitim rukama: kako vratiti, očistiti, rastaviti, promijeniti, veličinu, što učiniti ako rotor iskre i drugo

Ac220-230v Sidro Rotora Sidro Statora Zamijeniti Za Bosch Gbm500re Ručne Bušilice Rotor Električni Alati Za Popravak Rezervnih Dijelova Bosch Električni Alati > Alati / Proracun-Lako.cam

Kupiti Ac220-230v Sidro Sidro Rotora Sidra Za Udarni Bušilice Makita Hp1500 Rotor Električni Alat Za Popravak Rezervnih Dijelova / Alati - Proizvoda-Prodaja.cam

Zamjena i popravak sidra brusilice vlastitim rukama: kako vratiti, očistiti, rastaviti, promijeniti, veličinu, što učiniti ako rotor iskre i drugo
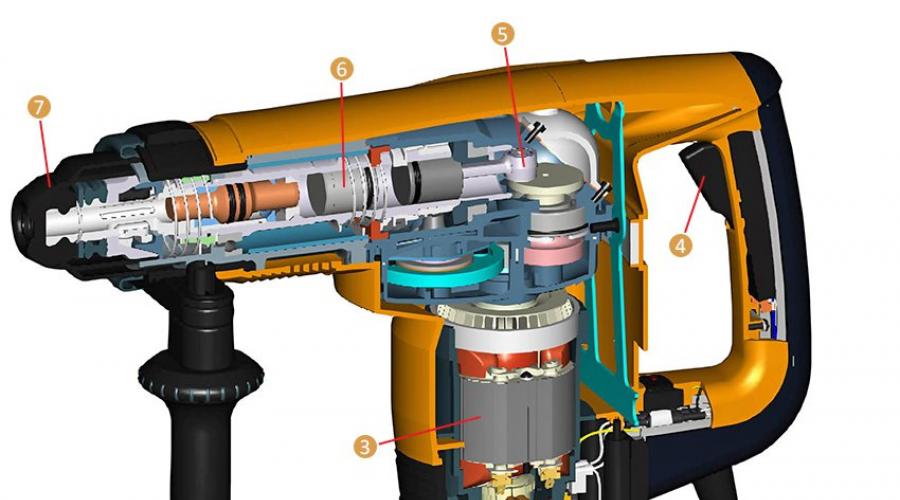
Popravite bušilicu za potočarku vlastitim rukama. Savjeti za popravak i rukovanje rupama sami. Kako se mijenja dugme za pokretanje
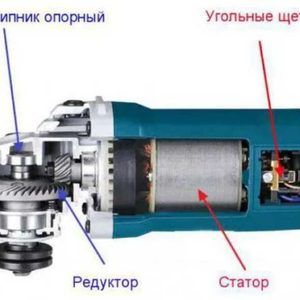
Popravak brusilice: utvrđivanje uzroka kvara i njihovo otklanjanje; uređaj i princip rada kutnih brusilica
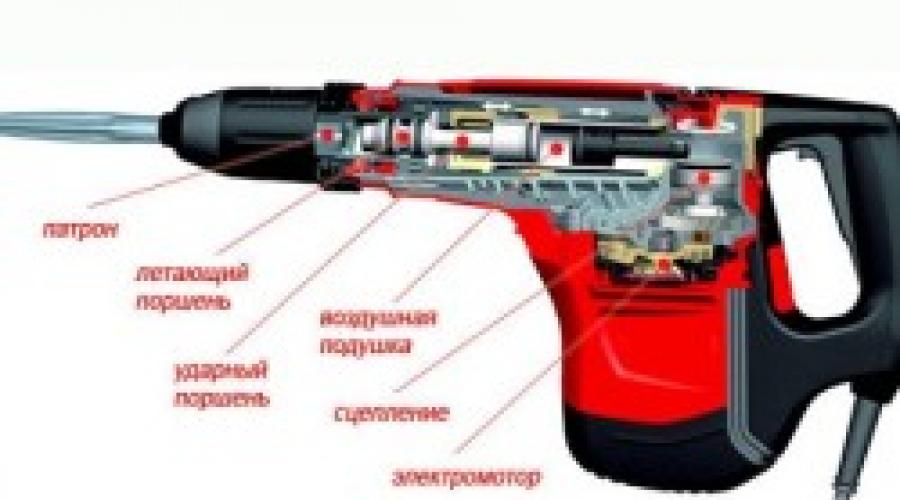
Popravak bajkalskog perforatora uradi sam. Samopopravak bušilice s čekićem. Dijagnostika žica i tipki bušilice s multimetrom

Kupi online Lanneret Standard 8~32 Mm Bočni Ključ Set Alata 1/2" Pogon Adapter Iščašenje Pretvarač Reduktor Električni šok Ključ Kombinirani Setovi | Pribor za ručni i električni alat / Global-Budget.news

Ac220-240v Električna Bušilica Sidro Rotor Prekidač Za Hitachi D10vc2 5 Zub Rotora Poslati Ugljika Zube Uređaja Dijelovi I Pribor < Alati \ www.us-coronavirus.news