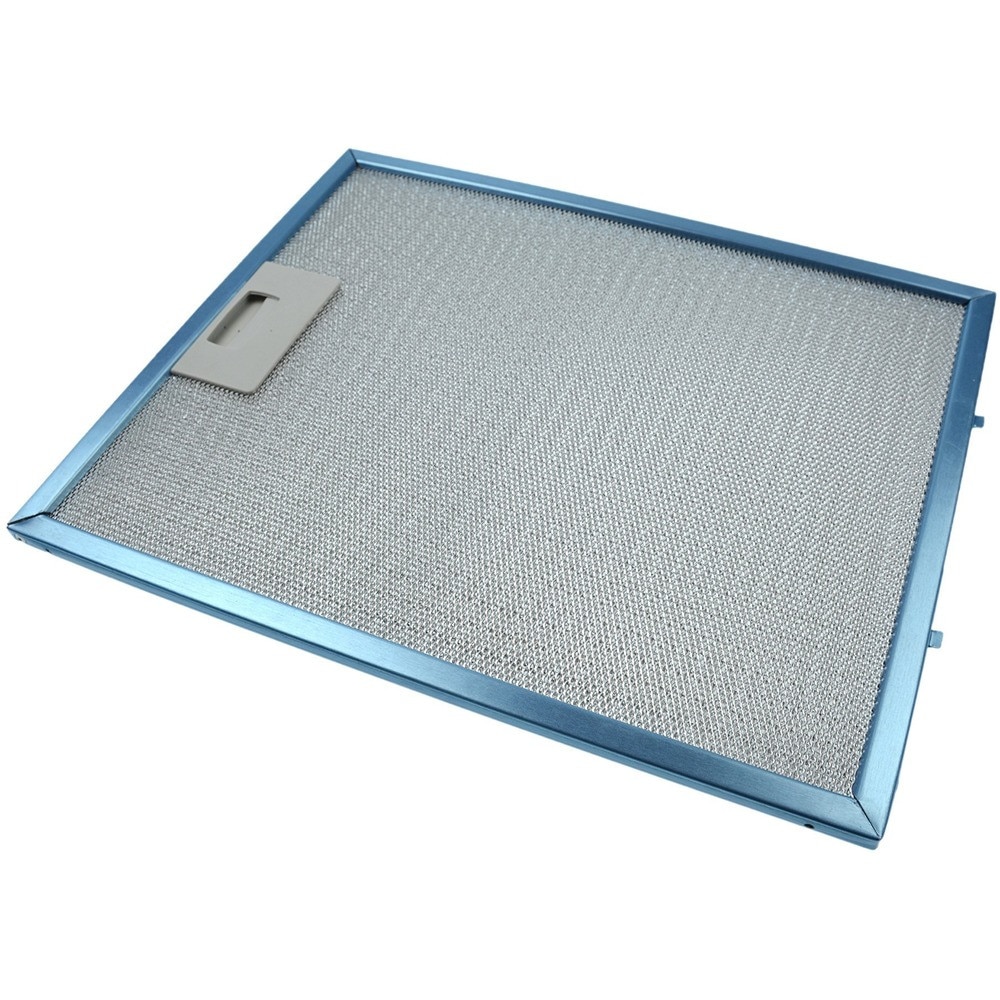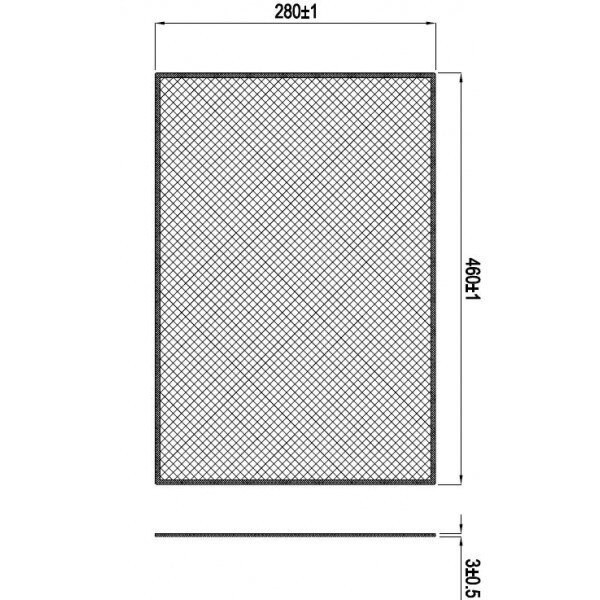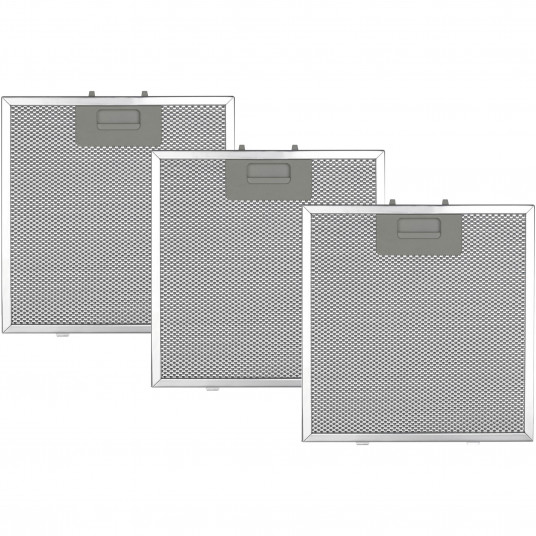
Imbunatatiti-va bucataria cu filtrul din aluminiu Star-Light ALF90HADT pentru hote decorative incorporate.

Filtru lavabil din aluminiu hota incorporabila Gorenje DF6116E, Whirlpool, Elica GF023C, GF023B, 4055103438, 50272636007, 4055364196,53.5x17.8x0.9 cm - eMAG.ro