BenBow Tornado Gun Premium Black | Valymo įrenginiai - Aukšto slėgio plovyklos, pramoniniai / statybiniai siurbliai | Įrankiai / Serviso įranga | AutoBaras.LT - viskas Jūsų automobiliui ir ne tik

Pirkti Lauko Ultralight Kastuvas, Sodo įrankiai Plieno Reikmenys, Sodo Pamišęs Mentele Gardenfarm Kastuvas Turistinius Mini C D5c1 ~ Kempingas & Žygiai > Industry4panevezys.lt

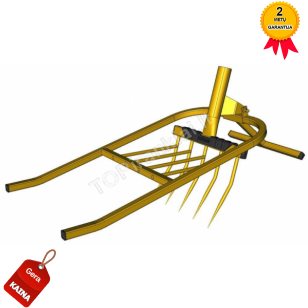
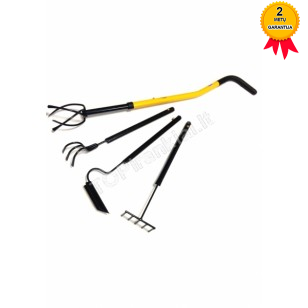
Averto - prekės daržui ir poilsiui - Kultivatorius „Tornadica“ https://averto.lt/lt/darzo-irankiai/sodo-irankiai -tornadica/rankinis-kultivatorius-tornado-tornadica/ | Facebook

BenBow Tornado Gun Premium Black | Valymo įrenginiai - Aukšto slėgio plovyklos, pramoniniai / statybiniai siurbliai | Įrankiai / Serviso įranga | AutoBaras.LT - viskas Jūsų automobiliui ir ne tik

Vejos traktorius Stiga Tornado 2098; 6,5 kW; mechaninė pavarų dėžė + Alyva - 2T0510481/ST1 - Vejos traktoriai - Žoliapjovės, vejapjovės

Stiga Tornado 7108e | Akumuliatoriniai | Traktoriukai | Prekių katalogas | STIGA, Husqvarna, Oleo-mac technika sodams ir parkams