Amazon.com: Compatible for Wireless Carplay Android Auto Car Stereo 6.2'' Android Car Radio with GPS, WiFi, SWC, BT, FM RDS, Mirror Link Compatible For Fiat Linea Punto EVO 2012-2015 Grande Linea 2007-2012 :

Amazon.com: RoverOne Car Stereo Bluetooth Radio GPS Navigation Head Unit for Fiat Abarth Punto EVO Linea 2012 2013 2014 2015 with Touch Screen Android USB MirrorLink WiFi : Electronics

Buy Custom fit car radio Fiat Punto Evo Android DAB Gps Bluetooth Wifi Usb Full Hd touchscreen display 6.2” Online at desertcartEcuador

Android 13 Auto radio For Fiat Linea Punto evo 2012-2015 Car Multimedia Bluetooth Touch Screen DVD Player Color WIFI Carplay - AliExpress

AMPrime 6.2'' 2din Android Car Radio For Fiat Punto 10-16/Linea 12-15 Car Multimedia Video Player Stereo GPS Navigation Carplay - AliExpress

Witson Android 12 Car Radio for FIAT Linea/ Punto 2012 2013 2014 2015 Auto GPS Navigation Car Multimedia Audio - China for FIAT Linea/ Punto 2012-2015, Car Multimedia | Made-in-China.com

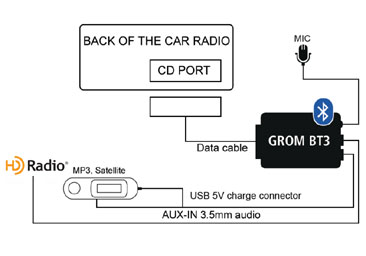
Yatour BTA Car Stereo Bluetooth MP3 Player for Alfa Romeo Fiat Punto Lancia Blaupunkt Radio Bluetooth Interface A2DP Music - AliExpress

Navigatie 1DIN cu Android Fiat Punto 1999 - 2005, 2GB RAM, Radio GPS Dual Zone, Display HD 9" Touchscreen reglabil 360 grade, Internet Wi-Fi, Bluetooth, MirrorLink, USB, Waze