Mikrofon előerősítő erősítő reverb tábla digitális kara ok surround késedelem etk3699 12v elektret dinamikus mikrofon, hangerősítő vásárlás online - Aktív összetevők | Elemeket-Bazar.cyou

AIYIMA Dinamikus Mikrofon Erősítő Testület Kondenzátor Mikrofon Forgalmazó Egy Bemeneti Hat Kimenet rendelés ~ pláza < Retail-Unique.cyou

CJ-9812 / MAX9812L alapú analóg mikrofon előerősítő modul (CJMCU-9812) - HESTORE - Elektronikai alkatrész kis- és nagykereskedelem

3,5 Mm-es Csatlakozó Headworn Fülhallgató Kondenzátor Mikrofon Mic Mike-ot A Számítógép, Tanári Beszéd, Pc, Laptop, Idegenvezető, Erősítő, Hangszóró rendelés \ Bolt | Vgihp.shop
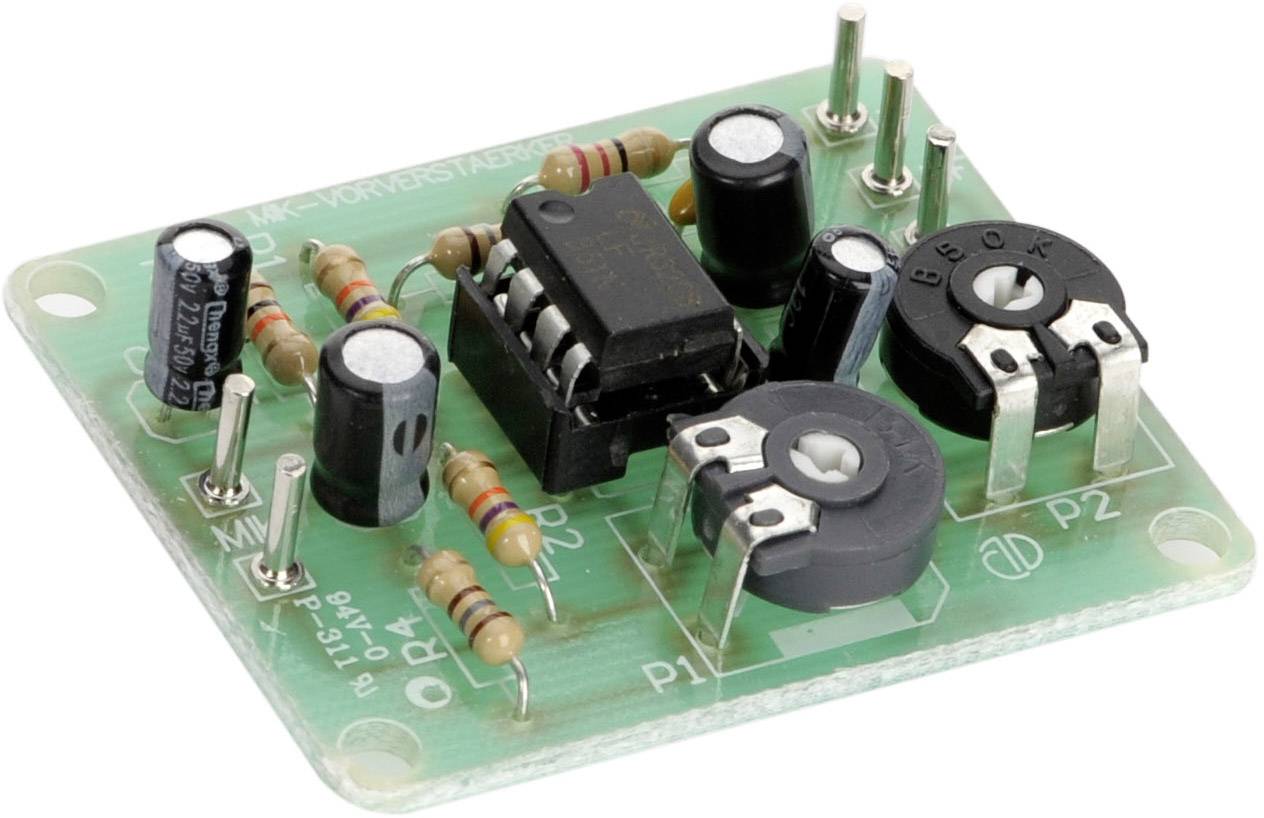
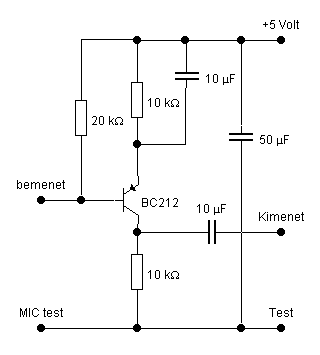
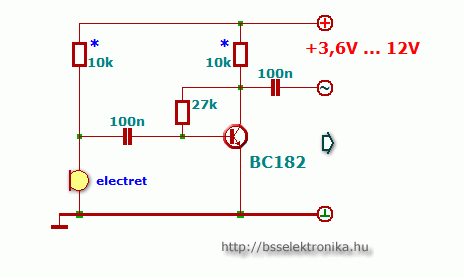
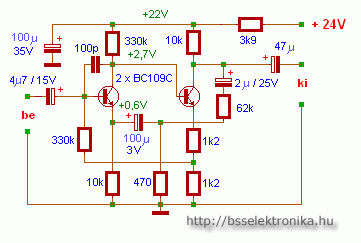
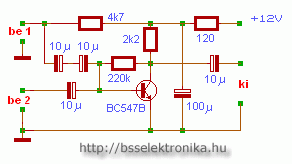
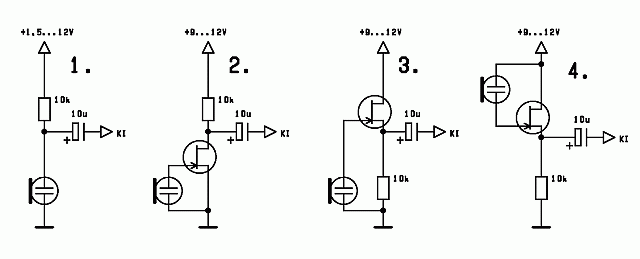
Egyszerű előerősítő, dinamikus mikrofonhoz - Hobbielektronika.hu - online elektronikai magazin és fórum

Kiváló Minőségű Csiptetős Számítógép Kondenzátor Mikrofon, 3,5 mm-es Sztereó Jack Microfone a Vezeték nélküli Rendszer, Mikrofon, Erősítő PC rendelés / Kedvezmények | Ertekesites-Raktar.today