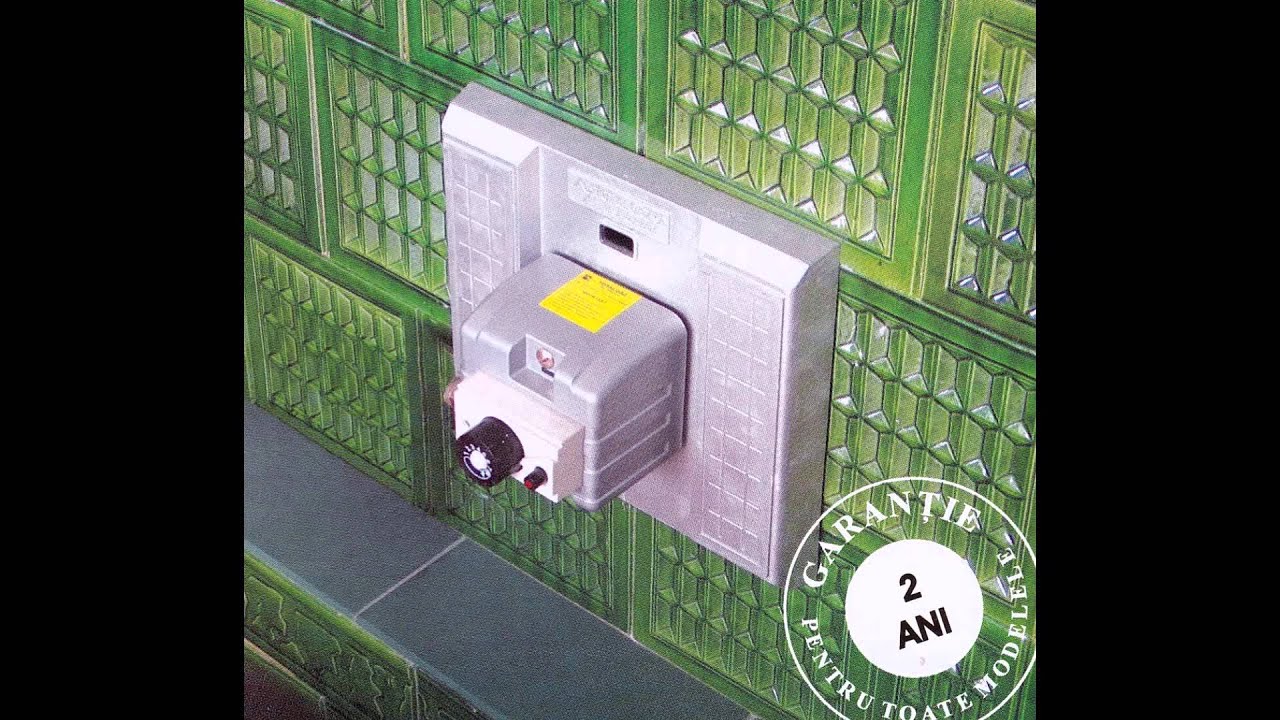
Arzator automat pentru sobe teracota cu economie gaz - Tg. Mures - Sc Impex Floris Srl, ID: 286252, pareri

Arzator automat pentru sobe teracota cu economie gaz - Tg. Mures - Sc Impex Floris Srl, ID: 286252, pareri

Arzator automat pentru sobe teracota cu economie gaz - Tg. Mures - Sc Impex Floris Srl, ID: 286252, pareri