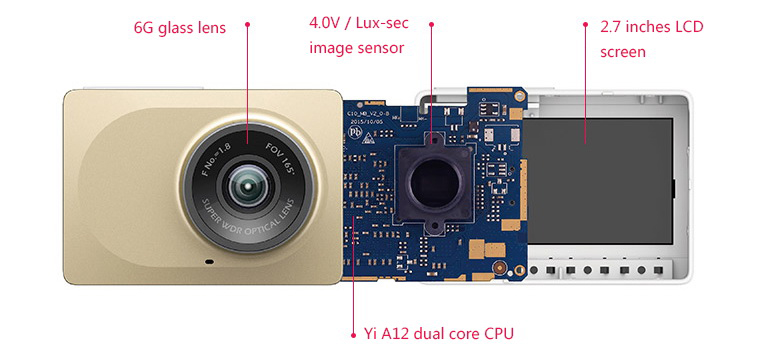
Original XiaoMi Yi 1080P Ambarella A7LS WIFI Sports Action Camera CN Version Sale, Price & Reviews | Gearbest

Original XiaoMi Yi 1080P Ambarella A7LS WIFI Sports Action Camera CN Version Sale, Price & Reviews | Gearbest

Original XiaoMi Yi 1080P Ambarella A7LS WIFI Sports Action Camera CN Version Sale, Price & Reviews | Gearbest

Original YI Data Cable 3.5M Long USB to Micro USB Cable Accessories for Yi Smart Dash Camera Xiaomi yi Car DVR|Data Cables| - AliExpress

Amazon.com : YI 4K Action and Sports Camera, 4K/30fps Video 12MP Raw Image with EIS, Live Stream, Voice Control - Black : Electronics

Mi Home Security Camera Wireless IP Surveillance System 1080p HD Night Vision (US Version with Warranty) - Xiaomi United States