ENLIGHTEN Ingenious Toys Military Carry Truck Vehicle Tank Army Tent & 7 Solders For War 308Pcs (811) - Ingenious Toys Military Carry Truck Vehicle Tank Army Tent & 7 Solders For War

TOYS 438pcs 6in3 Army Truck Helicopter Building Blocks For Children Military Soldiers gun Bricks | Wish

World War 2 WW 2 Military Armored Vehicle Transport Truck Building Blocks Bricks Sets Classic Model Kids Toys For Children Gift|Blocks| - AliExpress
Hot Stock】 WW2 Military Ambulance Army Truck Medic Soldiers Medical War Vehicle fit lego | Shopee Malaysia

Amazon.com: MZSmartTech Customized Heavy Missile Truck Army Vehicle Military Building Blocks for Adults (1377 PCS) : Toys & Games

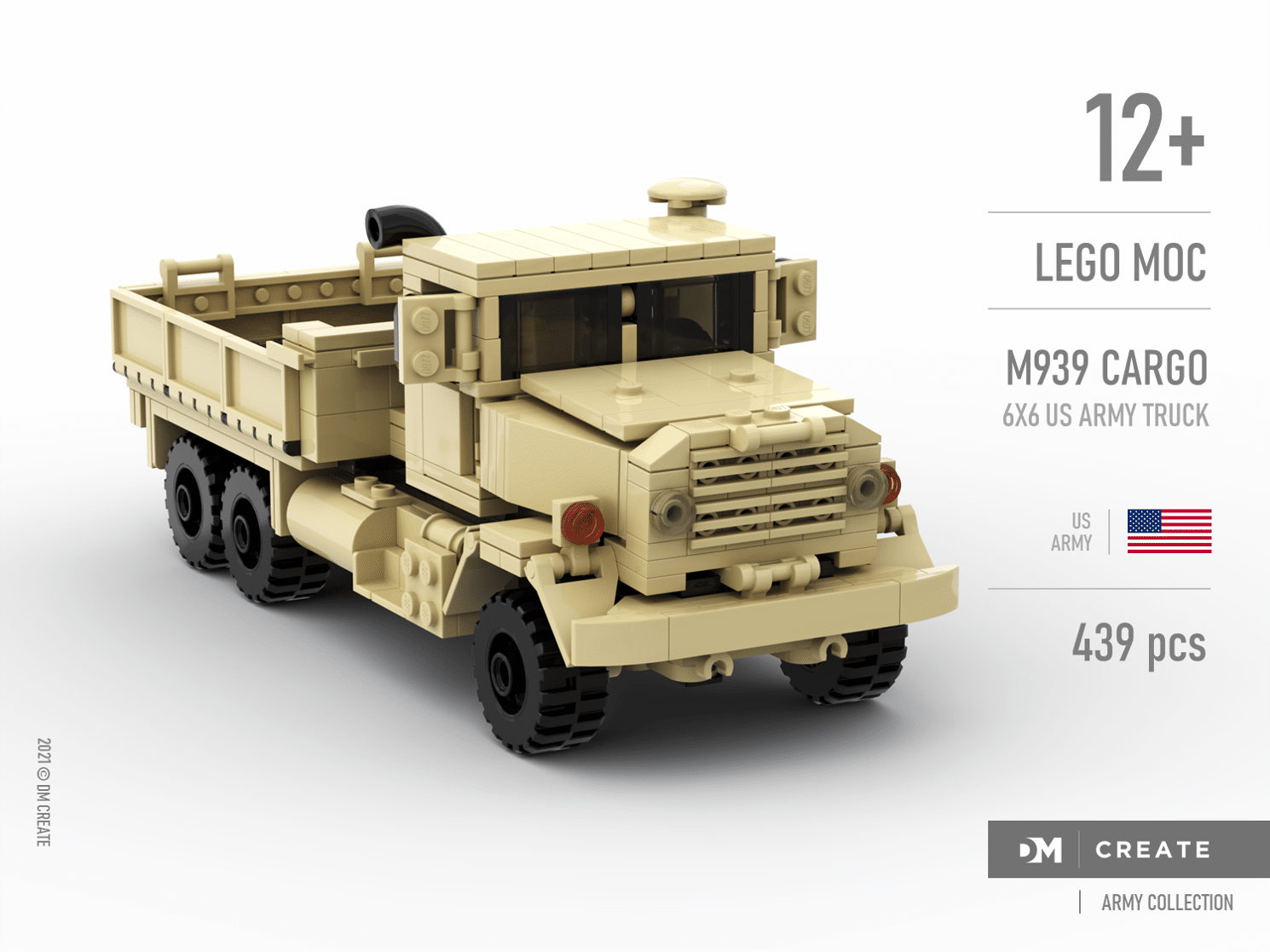
LEGO MOC Army collection - M939 Cargo - 6x6 US Army truck by DMcreate | Rebrickable - Build with LEGO