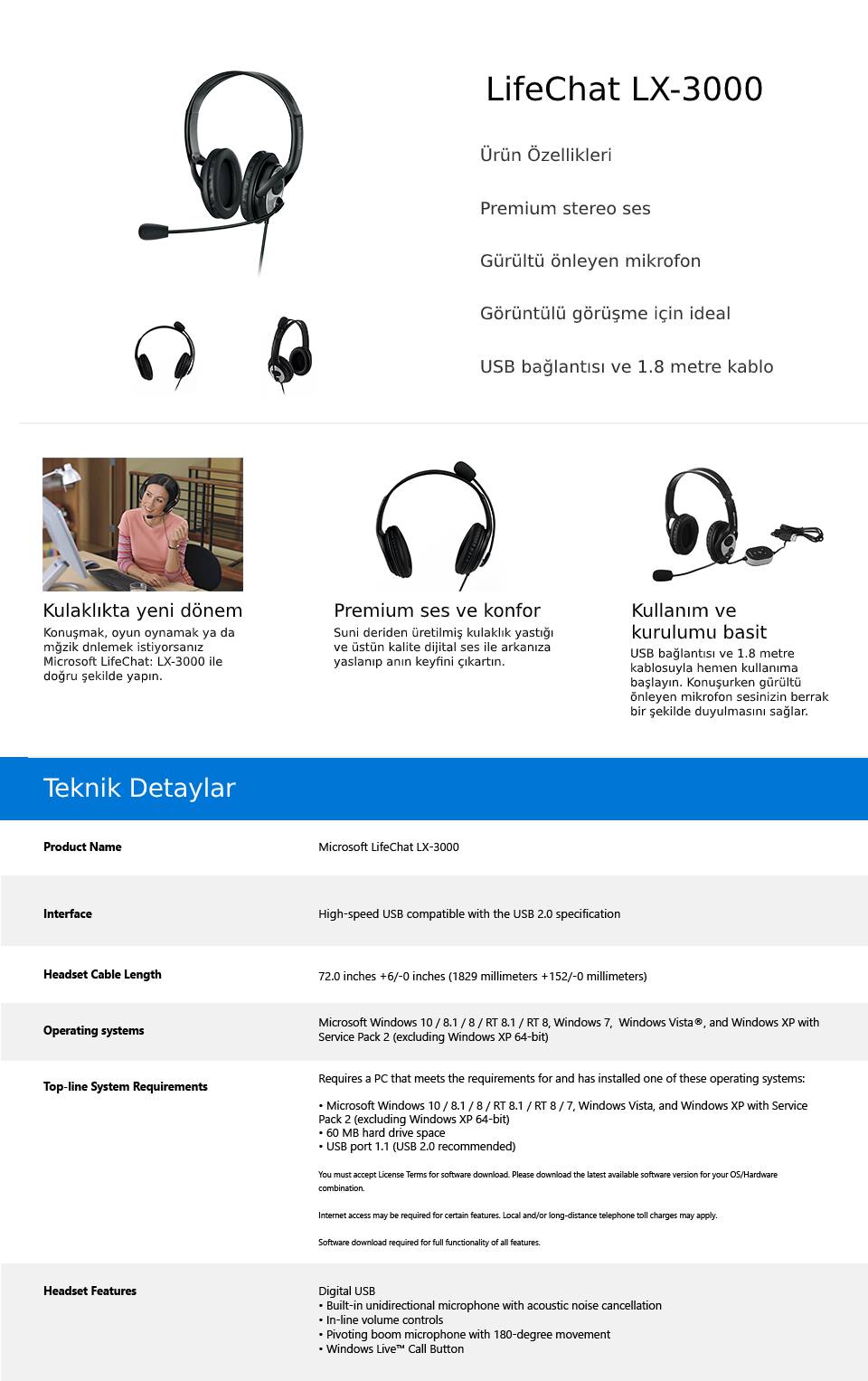
NullMini yedek kulak yastıkları Microsoft Lifechat LX3000 LX 3000 LX 3000 kulaklık kılıfı kulaklık kulaklık|Earphone Accessories| - AliExpress

kulaklık yastığı kadife Kulaklık Yastığı S Microsoft Lifechat Lx3000 Kulaklık Earpads Yedek Kulaklık Kulaklık Yastığı Pu Deri Sünger Köpük sipariş \ Taşınabilir ses ve video < Pazar-Taze.cam

Whiyo 1 çift Standart Yedek Kulak Pad Kulak Pedleri Yumuşak Yastık Microsoft Lifechat Lx 3000 Kulaklık satın almak online / Taşınabilir Ses Ve Video ~ Top-Items.cam

En Ucuz Microsoft LX-3000 LifeChat JUG-00014 L2 Mikrofonlu Kulak Üstü Kulaklık fiyatı incehesap.com 'da
![Yeni Microsoft LifeChat LX-3000 kulaklık [436830] | Bilgisayar Aksesuarları Toptan satış | merkandi.com.tr - Merkandi B2B Yeni Microsoft LifeChat LX-3000 kulaklık [436830] | Bilgisayar Aksesuarları Toptan satış | merkandi.com.tr - Merkandi B2B](https://merkandi.com.tr/images/offer/casque-micro-lx3000-1621329418-1621329583.jpeg)
Yeni Microsoft LifeChat LX-3000 kulaklık [436830] | Bilgisayar Aksesuarları Toptan satış | merkandi.com.tr - Merkandi B2B
![Yeni Microsoft LifeChat LX-3000 kulaklık [436830] | Bilgisayar Aksesuarları Toptan satış | merkandi.com.tr - Merkandi B2B Yeni Microsoft LifeChat LX-3000 kulaklık [436830] | Bilgisayar Aksesuarları Toptan satış | merkandi.com.tr - Merkandi B2B](https://merkandi.com.tr/images/offer/devant-casque-micro-microsoft-lx3000-1621329423-1621329583.jpeg)
Yeni Microsoft LifeChat LX-3000 kulaklık [436830] | Bilgisayar Aksesuarları Toptan satış | merkandi.com.tr - Merkandi B2B

1 çift kulak pedleri değiştirme yastık yastıkları köpük minder örtüsü bardak onarım bölümü Microsoft Lifechat LX 3000 kulaklık kulaklık|Earphone Accessories| - AliExpress

Whiyo 1 çift Standart Yedek Kulak Pad Kulak Pedleri Yumuşak Yastık Microsoft Lifechat Lx 3000 Kulaklık satın almak online / Taşınabilir Ses Ve Video ~ Top-Items.cam

NullMini yedek kulak yastıkları Microsoft Lifechat LX3000 LX 3000 LX 3000 kulaklık kulaklık kulaklık kılıfı kulaklık|Earphone Accessories| - AliExpress

Microsoft LifeChat LX-3000 Microfonlu Usb Kulaklık Mikrofonlu Kulaklık Microsoft - Herbirivar.com'da