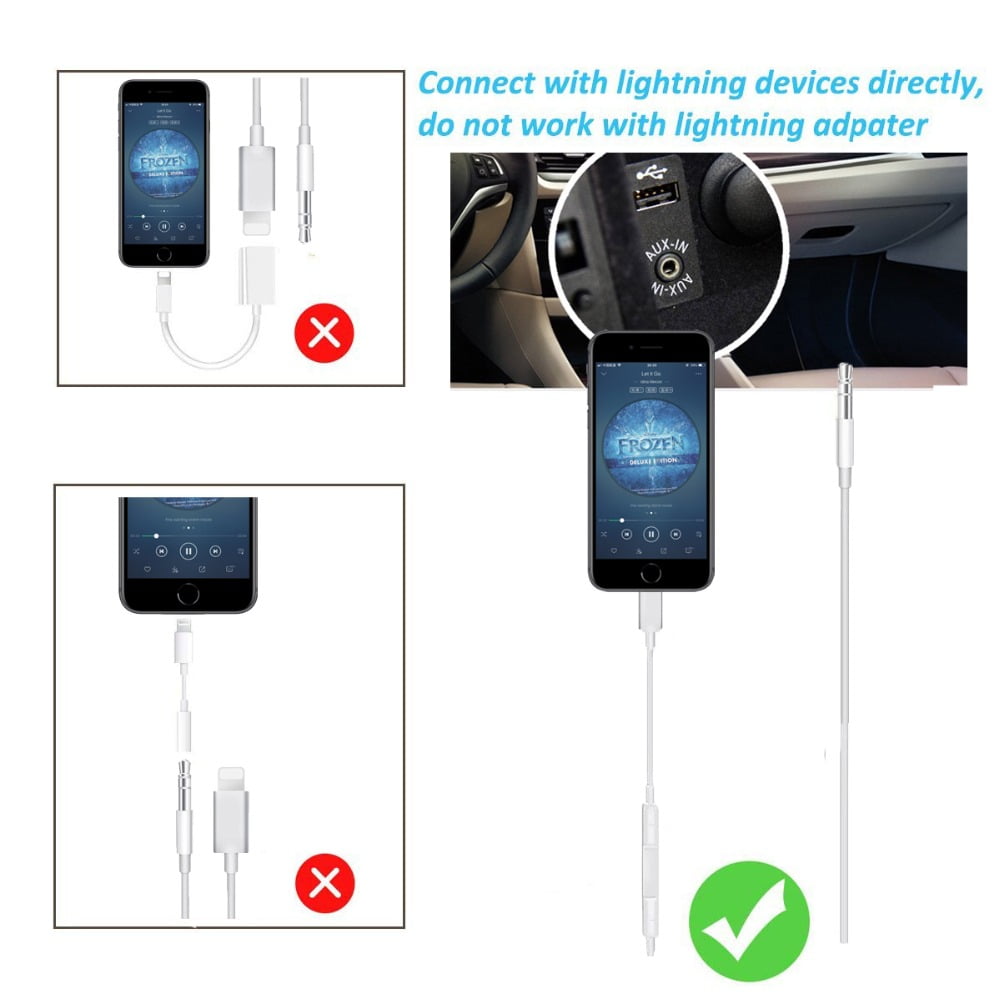
Car Aux Cable, Bluetooth connection, 3.5mm Jack to Lightning Headphone with Volume Control, for iPhone 7/8p/X/XSMAX/iPhone 11 Lightning Headphone to Car or Speaker cable - Walmart.com

Having issues skipping, stopping, and playing music tracks in your car over Bluetooth? Here's the fix! | iMore

GM G-MORE Bluetooth Receiver, Adapter Wireless Aux Receiver 4.1 Bluetooth Hands-free Car Kit Compatible With Iphone X IPhone 8 8 Plus 7 7plus Headsets Earbuds Car Audio Speaker Mini Home Stereo(Black) –

BC20 Bluetooth Car Kit, Supports AUX / Hands-free / Device Charging, for iPhone 6s & 6s Plus, iPhone 6 & 6 Plus, Samsung Galaxy S6 / S6 edge | Alexnld.com

Amazon.com: COMSOON Bluetooth Receiver with LCD Screen, Portable AUX Bluetooth 5.0 Adapter for Car/Home Stereo/Speaker/Wired Headphones, Dual Connection/Noise Cancelling/Hands-Free Calling/Music Streaming-Black : Electronics

Wireless Car Bluetooth AUX Audio Music Receiver Adapter Portable 3.5mm Streaming Car A2DP Wireless Bluetooth Receiver with Microphone for iPhone Samsung Android Cell Phones - Walmart.com

Yatour YTM07 Digital USB SD AUX Bluetooth ipod iphone interface for Smart 450 Lancia Lybra Fiat Brava Bravo Marea 8 Pin Plyer|interface inverter|interface transformationinterface diagram - AliExpress

Amazon.com: Bluetooth Transmitter/Receiver, 2-in-1 Wireless Bluetooth Adapter/Stereo Output/Just Need Connect to 3.5mm AUX Cord on The TV,Speaker,PC, iPhone, iPod, iPad, Tablets, MP3 Player Or Car : Electronics

Start Siri Wireless Bluetooth Car kit Handsfree 3.5mm AUX Audio Music Receiver Player Hands free Speaker 2.1A USB Car Charger rc|bluetooth car kit|wireless bluetooth car kitcar kit - AliExpress

Buy KUULAA Aux Bluetooth Adapter Dongle Cable For Car 3.5mm Jack Aux Bluetooth 5.0 4.2 4.0 Receiver Speaker Audio Music Transmitter at affordable prices — free shipping, real reviews with photos — Joom



:max_bytes(150000):strip_icc()/_hero_SQ_01LW1999388-1-4c0e27935edd4a5c9354b9e553826fce.jpg)








