Beginners Guide for Using Compression Stockings to Treat Varicose Veins | Blog | BASS Vein Center - BASS Vein Center

Compression Stockings For Varicose Veins. Compression Hosiery Royalty Free Cliparts, Vectors, And Stock Illustration. Image 60596098.

Medtrix Varicose Vein Stocking Thigh Support Premium Knee, Calf & Thigh Support - Buy Medtrix Varicose Vein Stocking Thigh Support Premium Knee, Calf & Thigh Support Online at Best Prices in India -

Medical Compression Stockings for Varicose Veins and Venouse Therapy. Compression Hosiery. Sock for Sports Isolated on White Back Stock Illustration - Illustration of disease, pregnancy: 180255799


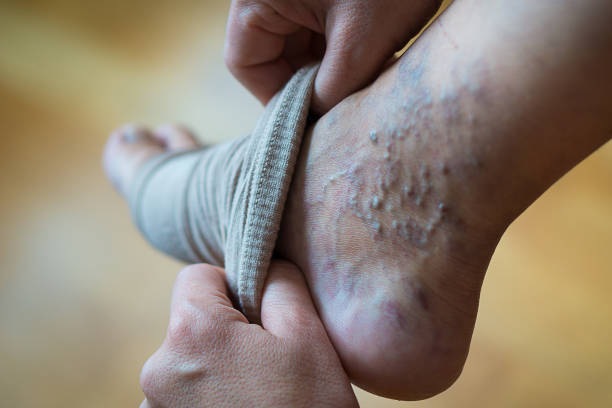
:max_bytes(150000):strip_icc()/Truform20-30mmHgCompressionStocking-c510ebb91d93441faf73681cd1ddc911.jpg)



:max_bytes(150000):strip_icc()/MojoSocksCompressionStockings-1ace2b0f89264eea9d1bf4c5785d38b7.jpg)