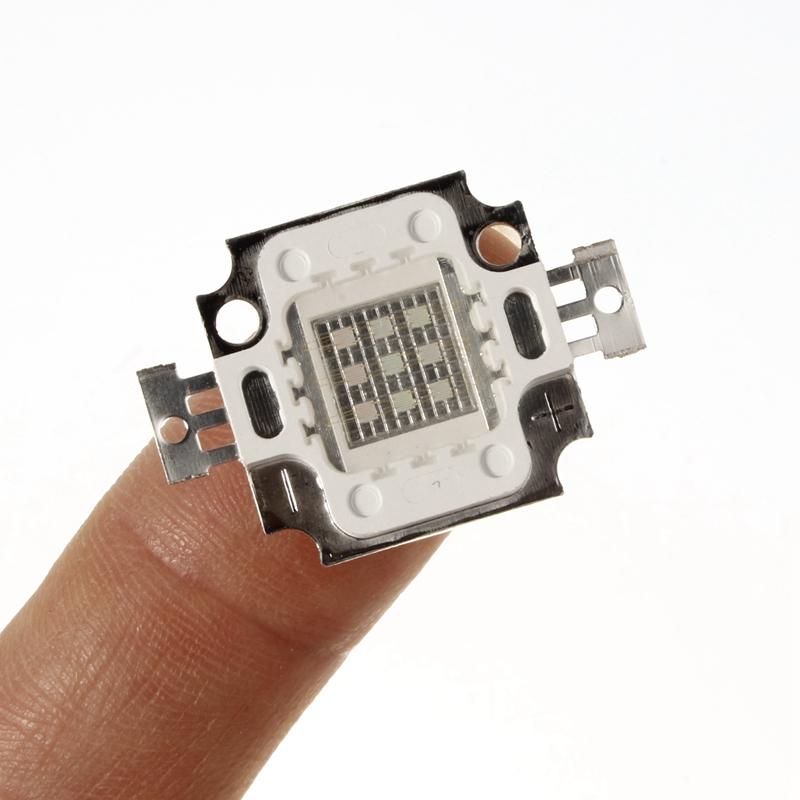
CHANZON High Power Led Chip 10W Purple Ultraviolet (UV 395nm / 900mA / DC 9V - 11V / 10 Watt) SMD COB Light Emitter Components Diode 10 W Ultra Violet Bulb Lamp Beads DIY Lighting - - Amazon.com

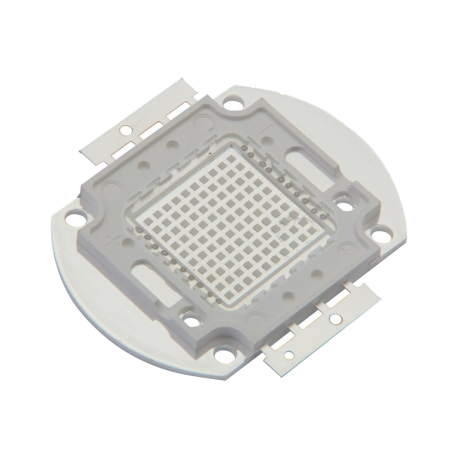
CHANZON High Power Led Chip 100W Purple Ultraviolet (UV 395nm / 3000mA / DC 30V - 34V / 100 Watt) SMD COB Light Emitter Components Diode 100 W Ultra Violet Bulb Lamp Beads DIY Lighting - - Amazon.com

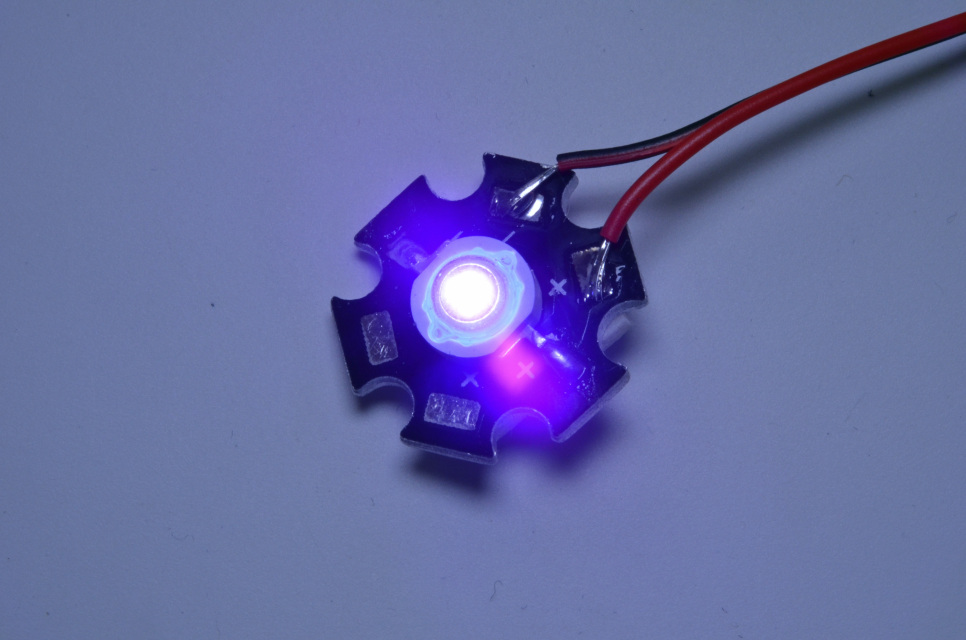
LEDGUHON 10Pcs High Power Pin LED Lamp Bead 3W UV 365nm 370nm Violet Light LED Chip UV Lamp Bead UV Led : Industrial & Scientific - Amazon.com

LED UV Black Light 150W LED Blacklight with Plug (10ft Power Cord) Ultraviolet Floodlight with Glow Tape Stage Lighting for Halloween Grow Party, Glow in The Dark, DJ Disco, Fluorescent Poster - -