
Amazon.com: LuDa 35A Brushless Electronic Controller with Switch for XLH Q901 Q902 Q903 : Toys & Games

Buy Generic Graupner DES 281 BBMG Torque Parkflyer 11.2mm Digital Servo JR Car RC Model Helicopter robot Online at Low Prices in India - Amazon.in

Amazon.com: FEETECH 35KG Servo Motor 7.4V High Voltage Waterproof High Torque RC Servo Full Metal Gear Digital Servo, Aluminium Case, Control Angle 180° for 1/8 1/10 RC Cars, TRAXXS x4, Baja, RC4WD,
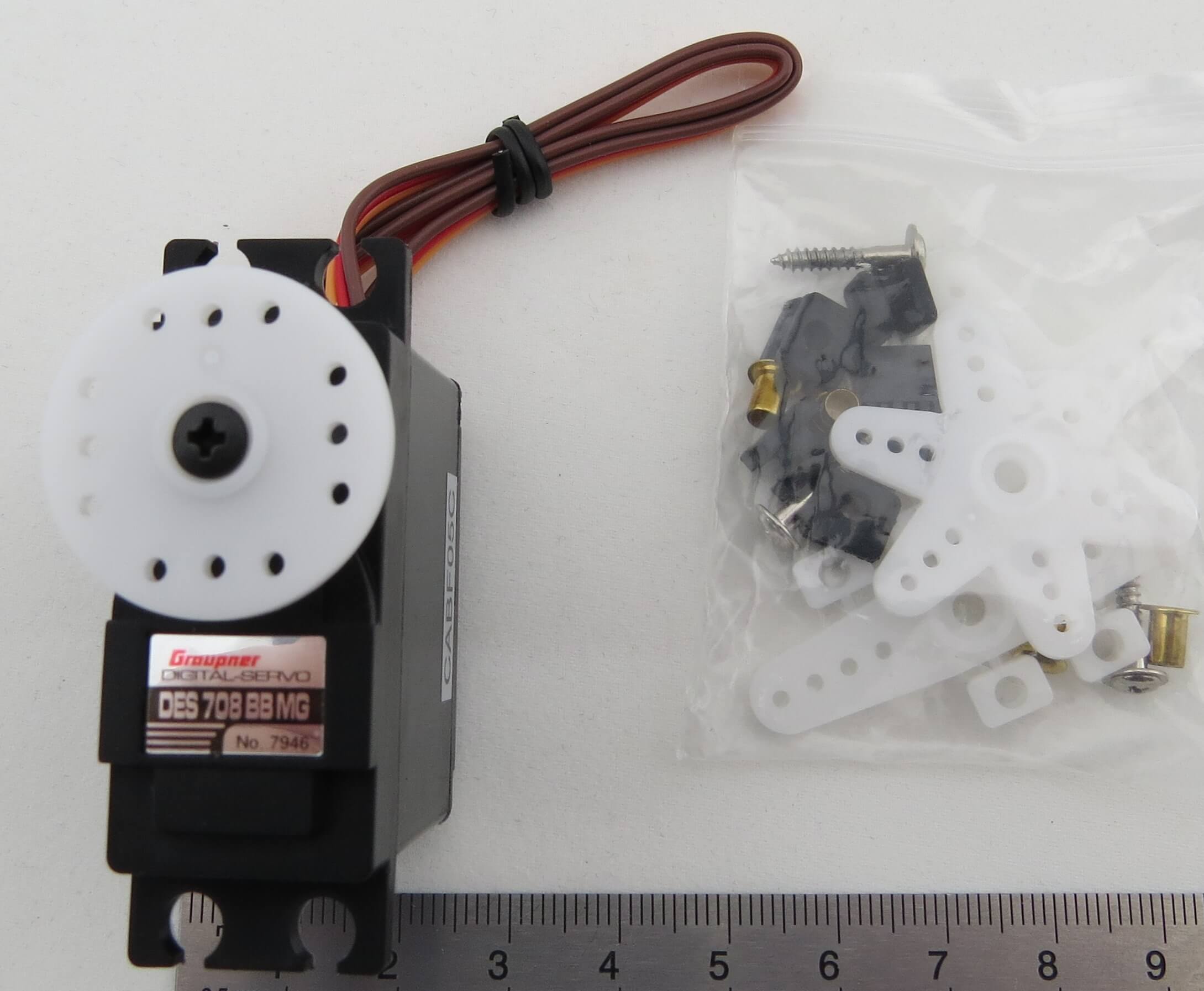
Servo DES-708 BB, MG, Digital.Graupner. With metal gear | Servos | RC articles | Fechtner model shop



















