Kína hallókészülék-beszállítók, gyártók, gyár - hallókészülékek nagykereskedelmi áron - EGÉSZSÉGÜGYI

Vásárlás Láthatatlan, Programozható, Digitális Hallókészülék Hordozható Mini Készülék Hallókészülékek Bal/jobb Fül Hang Erősítő S-13a Csepp Szállítás > Kedvezmény < www.lattelife.news

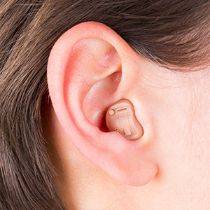
Feie Hallókészülék Audifonos Bte A Nagykereskedelmi ár A Magas Heairng Veszteség Siket Személy Szállítás Ingyenes S-303 kedvezmény < kedvezmény - Stock-Depot.cam

A 2020-as legjobb hallókészülékek értékelése (egészség) | Tippek a legjobb termékek és szolgáltatások kiválasztásához.

Vásárlás Láthatatlan, Programozható, Digitális Hallókészülék Hordozható Mini Készülék Hallókészülékek Bal/jobb Fül Hang Erősítő S-13a Csepp Szállítás > Kedvezmény < www.lattelife.news