Sarck Krabton - Dessin Streep - 43 x 43 x 75 cm | Dier | Cat activity centre, Cat activity, Cat scratcher

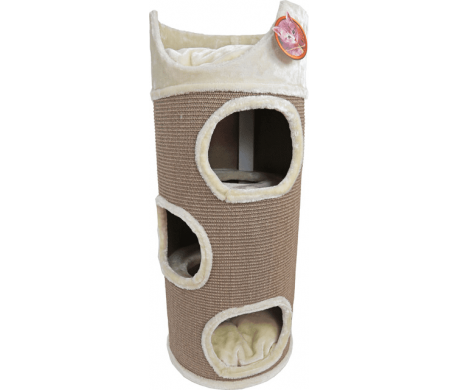
dibea KT00932 Cat Scratching Barrel, Scratching Tree for Cats, Height 100 cm, Choice of Colours- Buy Online in Angola at Desertcart - 81465657.

Cat Tree for Large Cats – Cat Mansion Light Grey – 71 inch 108 lbs 5 inch Ø poles – Total size 71x29x23 inch – Cat Scratcher scratching post activity center Cat