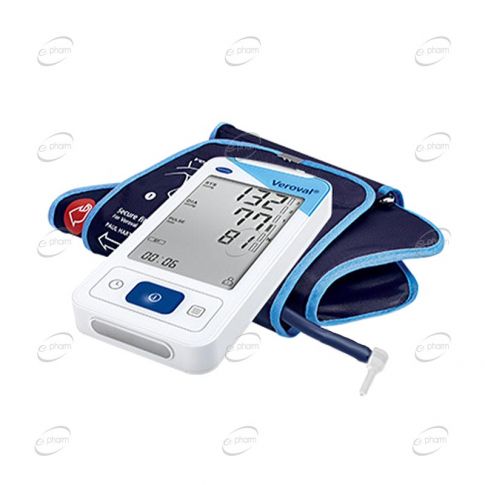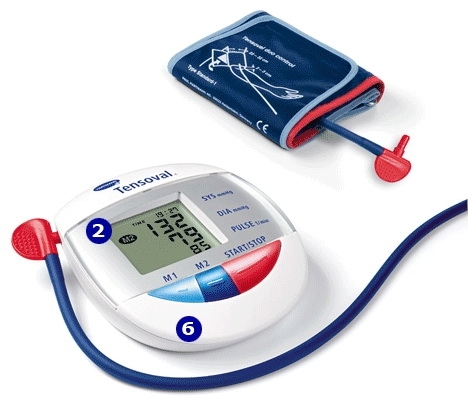
ХАРТМАН ЕЛЕКТРОНЕН АПАРАТ ЗА ИЗМЕРВАНЕ НА КРЪВНО НАЛЯГАНЕ ЗА ГОРНА ЧАСТ НА РЪКАТА размер 22 - 32 см ТЕНСОВАЛ ДУО КОНТРОЛ 900205 (UPPER ARM TYPE DIGITAL BLOOD PRESSURE MEASUREMENT MONITOR HARTMANN TENSOVAL
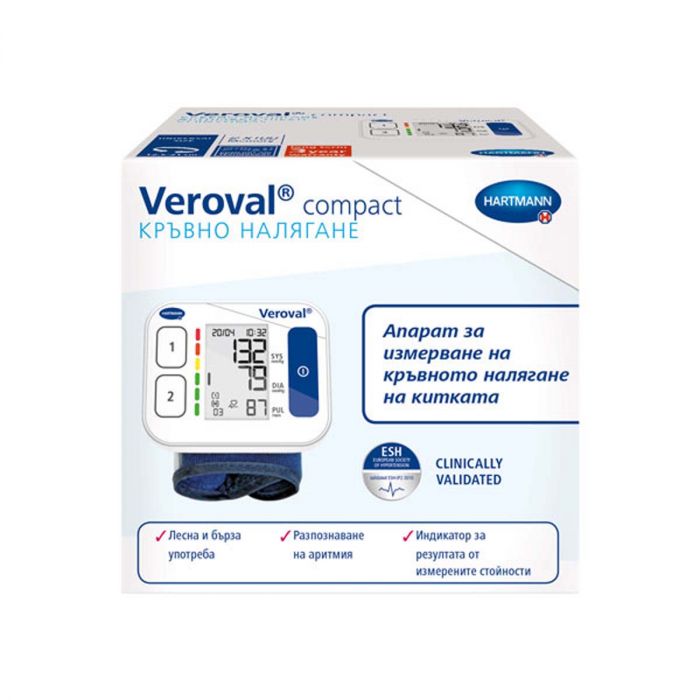
Електронен апарат за измерване на кръвно налягане за китка Hartmann Veroval Compact на изгодна цена | Galen.bg| Galen.bg
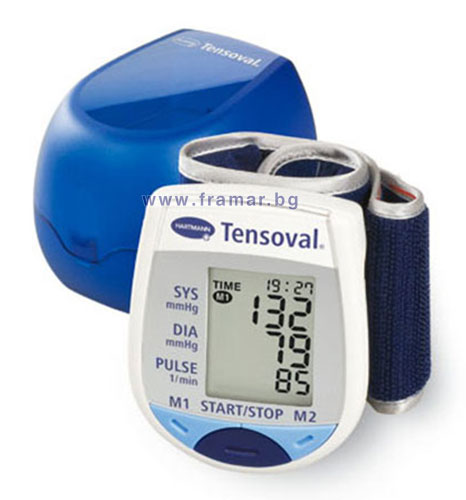
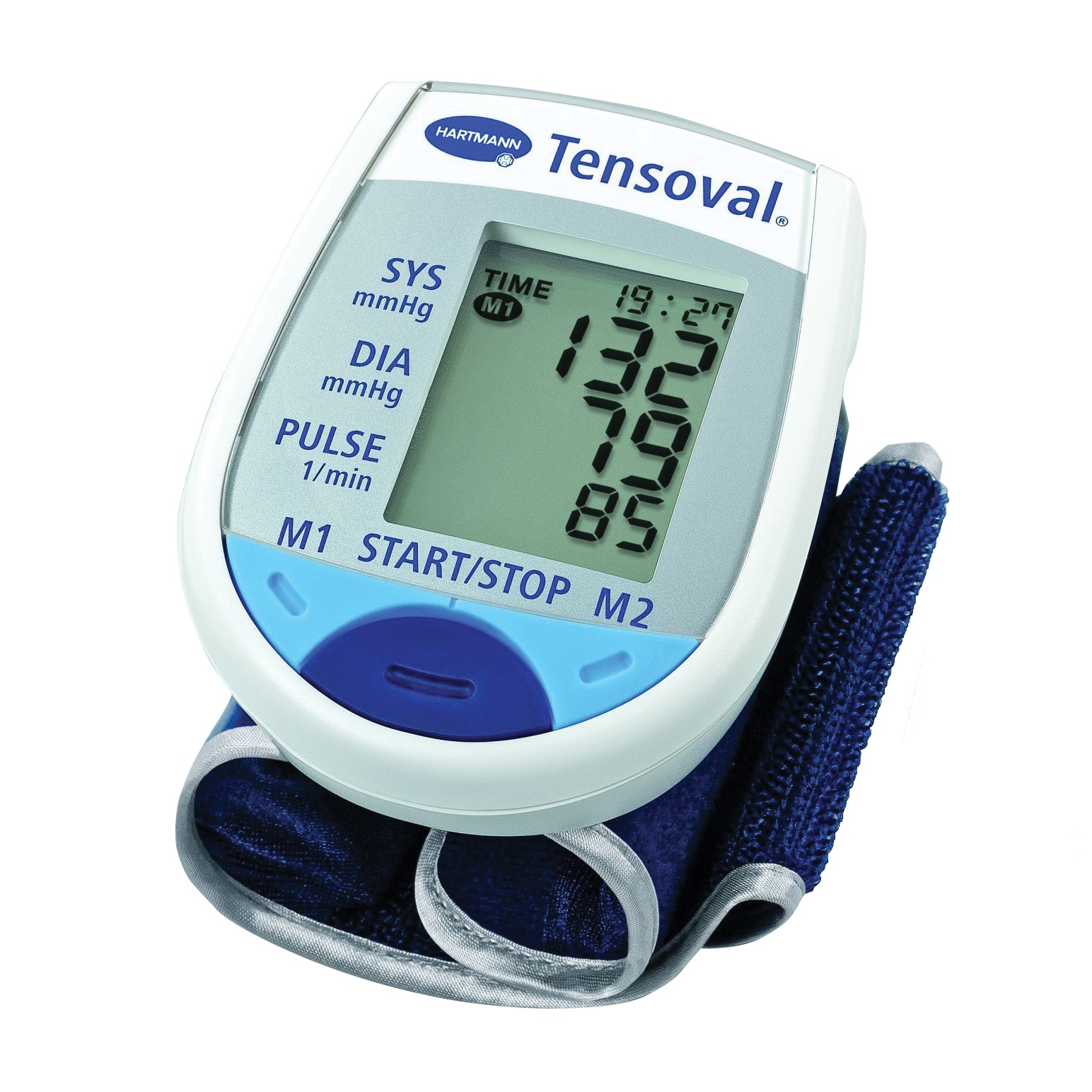
ХАРТМАН ЕЛЕКТРОНЕН АПАРАТ ЗА ИЗМЕРВАНЕ НА КРЪВНО НАЛЯГАНЕ НА КИТКА ТЕНСОВАЛ МОБИЛ 900115 (HARTMANN TENSOVAL MOBIL 900115 DIGITAL DEVICE FOR BLOOD PRESSURE MEASUREMENT WRIST TYPE), цена и информация
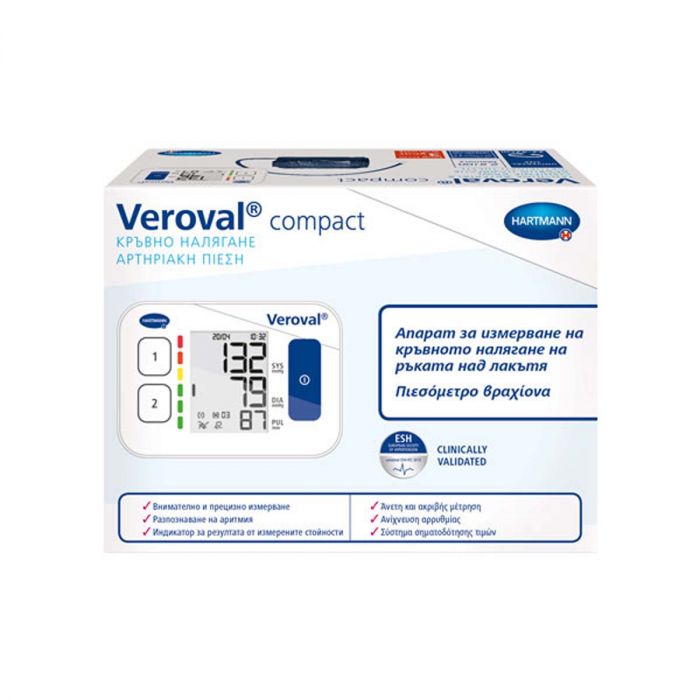
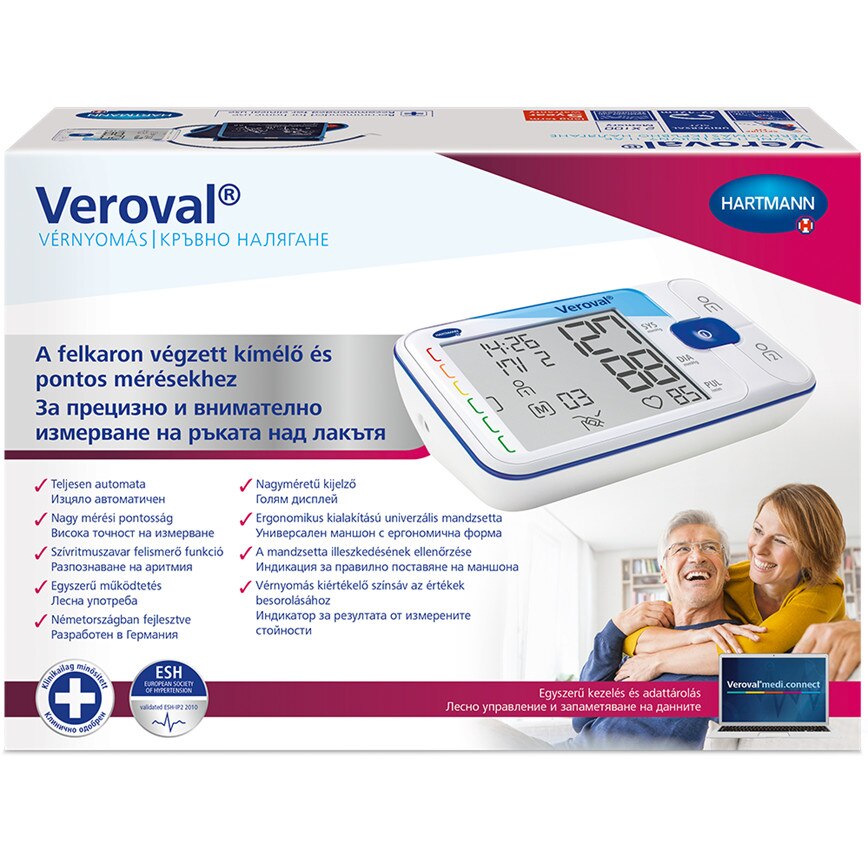
Електронен апарат за измерване на кръвно налягане за ръката над лакътя Hartmann Veroval Compact на изгодна цена | Galen.bg| Galen.bg
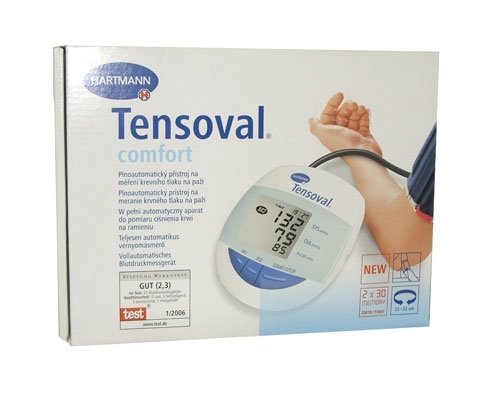
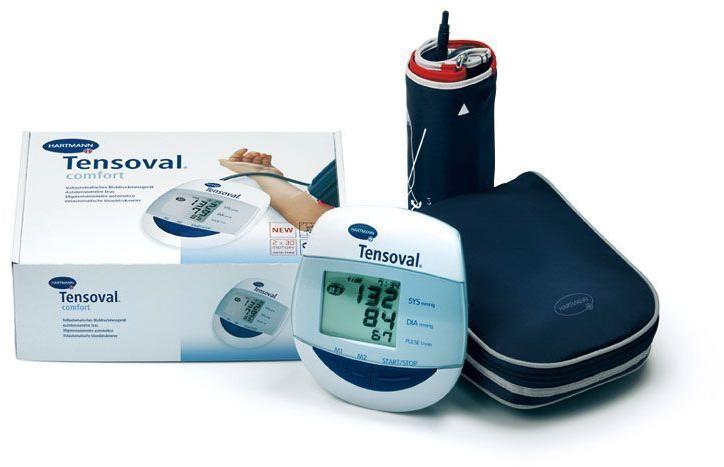
ХАРТМАН ЕЛЕКТРОНЕН АПАРАТ ЗА ИЗМЕРВАНЕ НА КРЪВНО НАЛЯГАНЕ ЗА ГОРНА ЧАСТ НА РЪКАТА ТЕНСОВАЛ КОМФОРТ 900173 (UPPER ARM TYPE DIGITAL BLOOD PRESSURE MEASUREMENT MONITOR HARTMANN TENSOVAL COMFORT WITH CUFF size 900173), цена

ХАРТМАН МАНШОН ЗА АПАРАТ ЗА КРЪВНО НАЛЯГАНЕ ТЕНСОВАЛ КОМФОРТ L РАЗМЕР / 32СМ Х 42СМ / - Безплатна доставка за цялата страна при поръчки над 39лв.
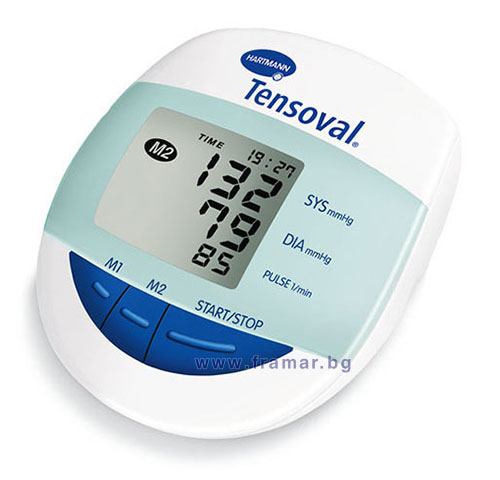
ХАРТМАН ЕЛЕКТРОНЕН АПАРАТ ЗА ИЗМЕРВАНЕ НА КРЪВНО НАЛЯГАНЕ ЗА ГОРНА ЧАСТ НА РЪКАТА ТЕНСОВАЛ КОМФОРТ 900173 (UPPER ARM TYPE DIGITAL BLOOD PRESSURE MEASUREMENT MONITOR HARTMANN TENSOVAL COMFORT WITH CUFF size 900173), цена