FAKTUM fali szekrény mikrohullámú sütő - Fagerland folt, antik, 60x92 cm (s09821117) - vélemények, ár összehasonlítás

Beépíthető mikrohullámú sütők - Fabuli Style Webáruház - Bútorkellékek és konyhai felszerelések boltja

Candy MIC 20 GDFBA beépíthető mikrohullámú sütő - árak, vásárlás, összehasonlítás - Áruház - Cserebirodalom

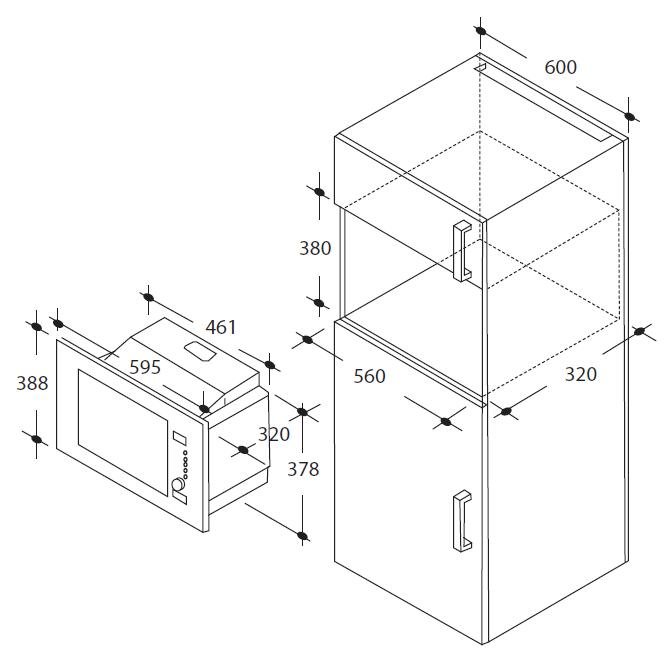
CANDY MIC 20 GDF BA × rusztikus beépíthető mikrohullámú sütő × Rusztikus beépíthető mikrohullámú sütő × parafalo.hu webáruház

ELADÓ ÚJ, Mikrohullámú sütő 34 liter 3200 W nyomógombos 2 magnetronos Ferrara-forni by Midea - Gasztroapró

CANDY MIC 20 GDF BA × rusztikus beépíthető mikrohullámú sütő × Rusztikus beépíthető mikrohullámú sütő × parafalo.hu webáruház