Vortex Crossfire HD 8x42 távcső, páramentes, tükröződésmentes, nagy tapadású, ütésálló, vízálló, 16 x 13 cm, 675 g, zöld - eMAG.hu

Vortex Crossfire HD 8x42 távcső, páramentes, tükröződésmentes, nagy tapadású, ütésálló, vízálló, 16 x 13 cm, 675 g, zöld - eMAG.hu

Vásárlás: Vortex Távcső - Árak összehasonlítása, Vortex Távcső boltok, olcsó ár, akciós Vortex Távcsövek

12 legjobb távcső - Online stílusú magazin a stílusról, a divatról, az etikettről, az életmódról és a legjobb termékek és szolgáltatások kiválasztásáról.

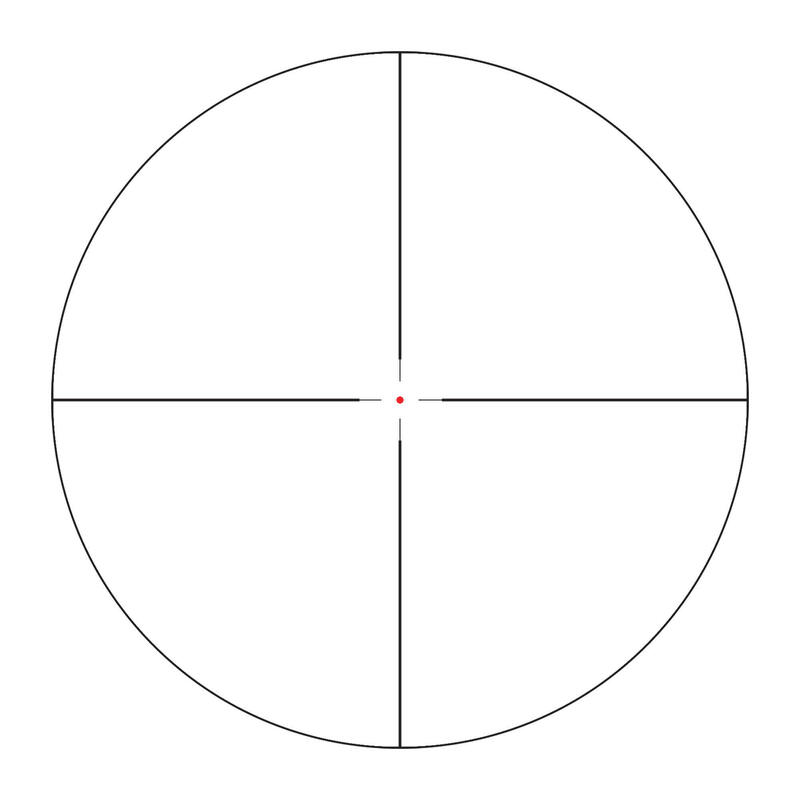
Vortex Crossfire II 3-12x56 AO V-Brite céltávcső - Proshooting.hu - ProShooting Kft fegyverbolt, Vadászat, vadászfegyverek, precíziós fegyverek, fegyvertisztítás, vadászbolt - Proshooting Budapesten