1 kos izolacijska vrečka iz netkane aluminijaste folije, izolirane vrečke za večkratno uporabo za nakupovanje, potovanje, piknik, armatura na vratih, hladilnik za hrano, vroča dostava - Temu Slovenia

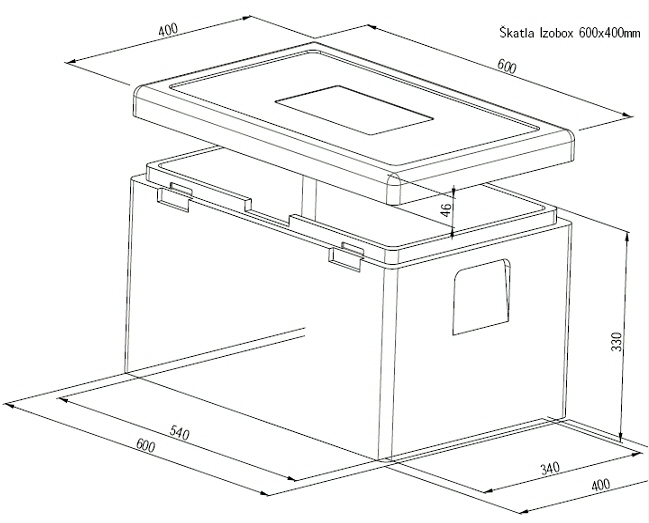
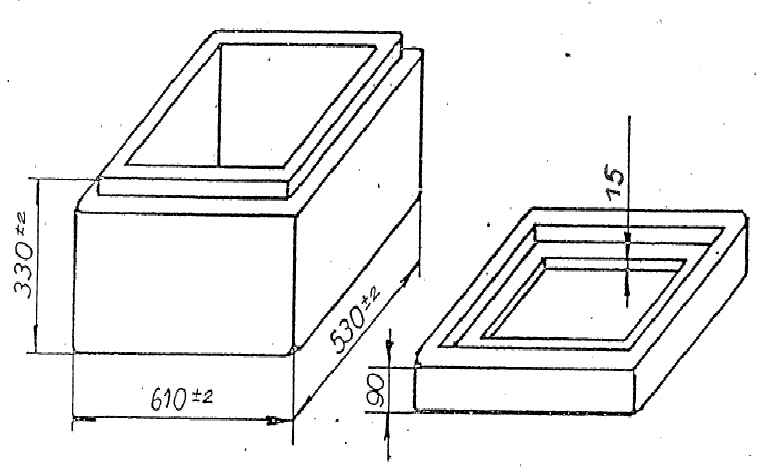
Termobox - za najenostavnejši transport hrane in živil v 2024 | OPREMA ZA CATERING IN DRUGI PRIPOMOČKI ZA POSTREŽBO | Horeca | Makaboshop.si | MAKABO d.o.o.

Termobox - za najenostavnejši transport hrane in živil v 2024 | OPREMA ZA CATERING IN DRUGI PRIPOMOČKI ZA POSTREŽBO | Horeca | Makaboshop.si | MAKABO d.o.o.

1 kos izolacijska vrečka iz netkane aluminijaste folije, izolirane vrečke za večkratno uporabo za nakupovanje, potovanje, piknik, armatura na vratih, hladilnik za hrano, vroča dostava - Temu Slovenia

Termobox - za najenostavnejši transport hrane in živil v 2024 | OPREMA ZA CATERING IN DRUGI PRIPOMOČKI ZA POSTREŽBO | Horeca | Makaboshop.si | MAKABO d.o.o.